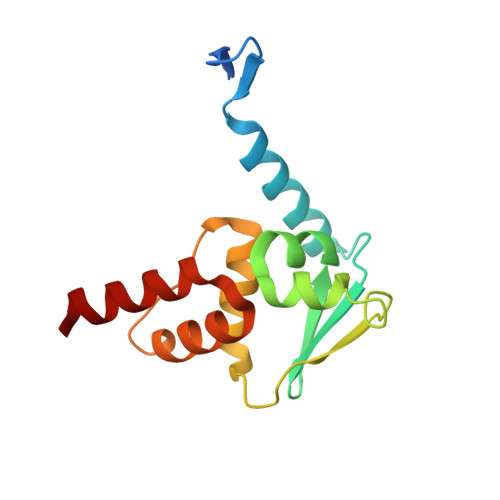
Discovery of high-affinity BCL6-binding peptide and its structure-activity relationship.
Sakamoto, K., Sogabe, S., Kamada, Y., Sakai, N., Asano, K., Yoshimatsu, M., Ida, K., Imaeda, Y., Sakamoto, J.I.(2017) Biochem Biophys Res Commun 482: 310-316
- PubMed: 27856253
- DOI: https://doi.org/10.1016/j.bbrc.2016.11.060
- Primary Citation of Related Structures:
5H7G, 5H7H - PubMed Abstract:
B cell lymphoma 6 (BCL6) is a transcriptional repressor that interacts with its corepressors BcoR and SMRT. Since this protein-protein interaction (PPI) induces activation and differentiation of B lymphocytes, BCL6 has been an attractive drug target for potential autoimmune disease treatments. Here we report a novel BCL6 inhibitory peptide, F1324 (Ac-LWYTDIRMSWRVP-OH), which we discovered using phage display technology; we also discuss this peptide's structure-activity relationship (SAR). For BCL6(5-129) binding, K D and IC 50 values of F1324 were 0.57 nM and 1 nM according to the results of an SPR analysis and cell-free ELISA assay, respectively. In contrast, BcoR(Arg498-514Pro) and SMRT(Leu1422-Arg1438) exhibited relatively weak micromole-order binding to BCL6. Furthermore, Fusion protein AcGFP-F1324 transiently expressed in HEK293T cells inhibited intracellular PPI in cell-based M2H assay. By examination of the truncation and fragmentation of F1324, the C-terminal sequence WRVP, which is similar to the BcoR(509-512) sequence WVVP, was identified as being critical for BCL6 binding. In addition, subsequent single-crystal X-ray diffraction analysis of F1324/BCL6(5-129) complex revealed that the high affinity of F1324 was caused by effective interaction of its side chains while its main chain structure was similar to that of BcoR(Arg498-514Pro). To our knowledge, F1324 is the strongest BCL6-binding peptide yet reported.
- Pharmaceutical Research Division, Takeda Pharmaceutical Company Limited, 26-1, Muraoka-Higashi 2-chome, Fujisawa, Kanagawa 251-8555, Japan. Electronic address: koutarou.sakamoto@takeda.com.
Organizational Affiliation: