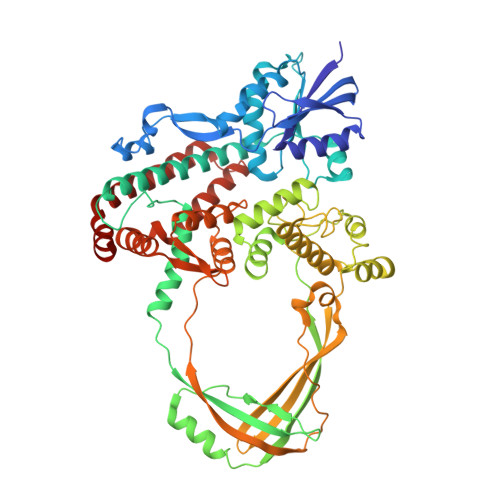
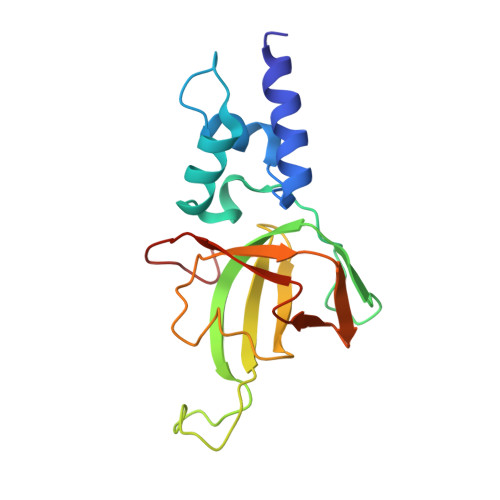
Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Goto-Ito, S., Yamagata, A., Takahashi, T.S., Sato, Y., Fukai, S.(2017) Sci Rep 7: 42123-42123
- PubMed: 28176834
- DOI: https://doi.org/10.1038/srep42123
- Primary Citation of Related Structures:
5GVC, 5GVD, 5GVE - PubMed Abstract:
Topoisomerase IIIβ (TOP3β) is a DNA/RNA topoisomerase that has been implicated in epigenetic or translational control of gene expression. In cells, TOP3β co-exists with its specific auxiliary factor, TDRD3. TDRD3 serves as a scaffold protein to recruit TOP3β to its DNA/RNA substrates accumulating in specific cellular sites such as methylated chromatins or neural stress granules. Here we report the crystal structures of the catalytic domain of TOP3β, the DUF1767-OB-fold domains of TDRD3 and their complex at 3.44 Å, 1.62 Å and 3.6 Å resolutions, respectively. The toroidal-shaped catalytic domain of TOP3β binds the OB-fold domain of TDRD3. The TDRD3 OB-fold domain harbors the insertion loop, which is protruding from the core structure. Both the insertion loop and core region interact with TOP3β. Our pull-down binding assays showed that hydrophobic characters of the core surface and the amino- and carboxy-terminal regions of the insertion loop are essential for the interaction. Furthermore, by comparison with the structure of the homologous Topoisomerase IIIα (TOP3α)-RMI1 complex, we identified Arg96, Val109, Phe139 and the short insertion loop of TDRD3 as the critical structural elements for the specific interaction with TOP3β to avoid the non-cognate interaction with TOP3α.
- Structural Biology Laboratory, Structural Life Science Division, Synchrotron Radiation Research Organization and Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo 113-0032, Japan.
Organizational Affiliation: