Structural analyses of the nucleosome complexes with human testis-specific histone variants, hTh2a and hTh2b
Padavattan, S., Thiruselvam, V., Shinagawa, T., Hasegawa, K., Kumasaka, T., Ishii, S., Kumarevel, T.(2017) Biophys Chem 221: 41-48
- PubMed: 27992841
- DOI: https://doi.org/10.1016/j.bpc.2016.11.013
- Primary Citation of Related Structures:
5GSU, 5GT0, 5GT3 - PubMed Abstract:
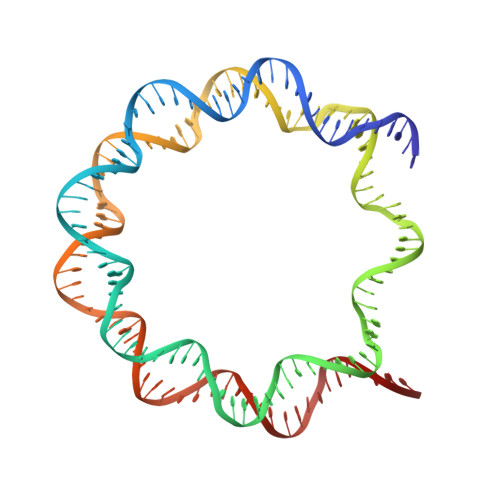
Th2a and Th2b are the testis-specific histone variants highly expressed during spermatogenesis. Approximately 4% of the genome is retained in nucleosomes in mature human sperm, which is enriched at loci of developmental importance. Our recent studies revealed that the mouse histone variant homologs TH2a and TH2b are involved in reprogramming. In the present work, we report three nucleosome structures (NCPs) with human testis-specific histone variants hTh2a and hTh2b, [hGcH (hTh2a-hTh2b-H3-H4), hGcHV1 (hTh2a-H2b-H3-H4) and hGcHV2 (H2a-hTh2b-H3-H4)] and a 146-base pair (bp) duplex DNA fragment at ~3.0Å resolutions. These crystal structures revealed two major changes within the nucleosomes, either with hTh2a, hTh2b or both variants, as compared to the canonical counterpart. First, the H-bonding interactions between the L1-L1' interfaces mediated by the hTh2a/hTh2a' L1-loops are lost. Second, the histone dimer-DNA contacts are considerably reduced, and these changes are localized around ±31 to 35-bp from the nucleosome entry/exit sites. Thus, the modified functional residues at the N- and C-terminal ends of histone variants are responsible for the observed structural changes and regulate the gene expression through specific structural alterations in the chromatin by modulating the chromatin-associated binding proteins.
- RIKEN SPring-8 Center, Harima Institute, 1-1-1 Kouto, Sayo, Hyogo 679-5148, Japan.
Organizational Affiliation: