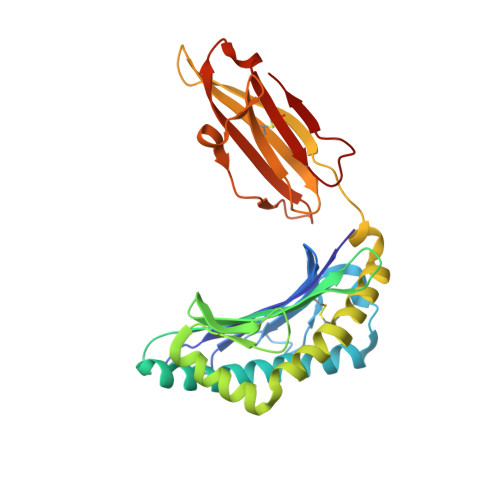
Dual non-contiguous peptide occupancy of HLA class I evoke antiviral human CD8 T cell response and form neo-epitopes with self-antigens
Xiao, Z., Ye, Z., Tadwal, V.S., Shen, M., Ren, E.C.(2017) Sci Rep 7: 5072-5072
- PubMed: 28698575
- DOI: https://doi.org/10.1038/s41598-017-05171-w
- Primary Citation of Related Structures:
5GRD, 5GRG, 5GSD - PubMed Abstract:
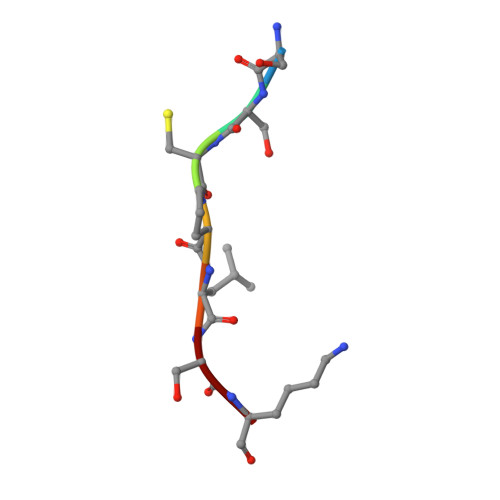
Host CD8 T cell response to viral infections involves recognition of 8-10-mer peptides presented by MHC-I molecules. However, proteasomes generate predominantly 2-7-mer peptides, but the role of these peptides is largely unknown. Here, we show that single short peptides of <8-mer from Latent Membrane Protein 2 (LMP2) of Epstein Barr Virus (EBV) can bind HLA-A*11:01 and stimulate CD8 + cells. Surprisingly, two peptide fragments between 4-7-mer derived from LMP2 (340-349) were able to complement each other, forming combination epitopes that can stimulate specific CD8 + T cell responses. Moreover, peptides from self-antigens can complement non-self peptides within the HLA binding cleft, forming neoepitopes. Solved structures of a tetra-complex comprising two peptides, HLA and β2-microglobulin revealed the free terminals of the two peptides to adopt an upward conformation directed towards the T cell receptor. Our results demonstrate a previously unknown mix-and-match combination of dual peptide occupancy in HLA that can generate vast combinatorial complexity.
- Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), 8A Biomedical Grove, #03-06 Immunos, Singapore, 138648, Singapore.
Organizational Affiliation: