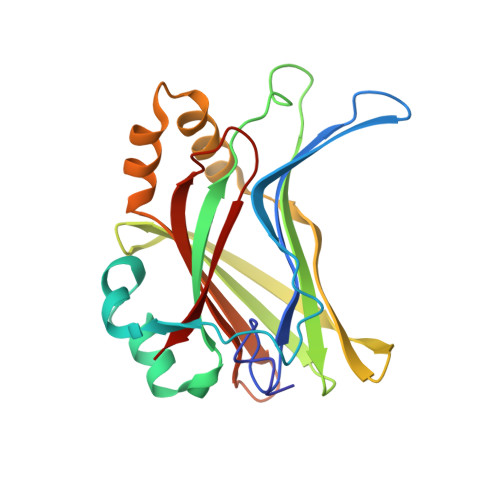
Crystal structure of TAZ-TEAD complex reveals a distinct interaction mode from that of YAP-TEAD complex
Kaan, H.Y.K., Chan, S.W., Tan, S.K.J., Guo, F., Lim, C.J., Hong, W., Song, H.(2017) Sci Rep 7: 2035-2035
- PubMed: 28515457
- DOI: https://doi.org/10.1038/s41598-017-02219-9
- Primary Citation of Related Structures:
5GN0 - PubMed Abstract:
The Hippo pathway is a tumor suppressor pathway that is implicated in the regulation of organ size. The pathway has three components: the upstream regulatory factors, the kinase core, and the downstream transcriptional machinery, which consists of YAP, TAZ (transcription co-activators) and TEAD (transcription factor). Formation of YAP/TAZ-TEAD complexes leads to the transcription of growth-promoting genes. Herein, we report the crystal structure of TAZ-TEAD4 complex, which reveals two binding modes. The first is similar to the published YAP-TEAD structure. The second is a unique binding mode, whereby two molecules of TAZ bind to and bridge two molecules of TEAD4. We validated the latter using cross-linking and multi-angle light scattering. Using siRNA, we showed that TAZ knockdown leads to a decrease in TEAD4 dimerization. Lastly, results from luciferase assays, using YAP/TAZ transfected or knockdown cells, give support to the non-redundancy of YAP/TAZ co-activators in regulating gene expression in the Hippo pathway.
- Institute of Molecular and Cell Biology, A*STAR (Agency for Science, Technology and Resesarch), 61 Biopolis Drive, Singapore, 138673, Singapore.
Organizational Affiliation: