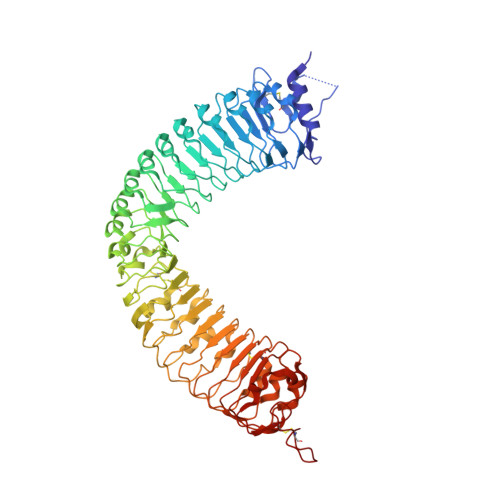
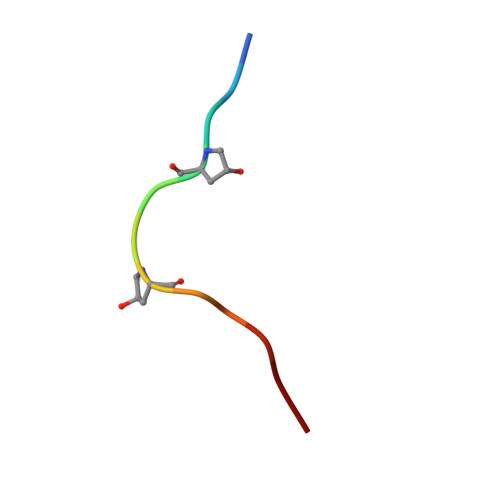
Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide
Morita, J., Kato, K., Nakane, T., Kondo, Y., Fukuda, H., Nishimasu, H., Ishitani, R., Nureki, O.(2016) Nat Commun 7: 12383-12383
- PubMed: 27498761
- DOI: https://doi.org/10.1038/ncomms12383
- Primary Citation of Related Structures:
5GIJ - PubMed Abstract:
In plants, leucine-rich repeat receptor-like kinases (LRR-RKs) perceive ligands, including peptides and small molecules, to regulate various physiological processes. TDIF, a member of the CLE peptide family, specifically interacts with the LRR-RK TDR to inhibit meristem differentiation into tracheary elements, and promotes cell proliferation. Here we report the crystal structure of the extracellular domain of TDR in complex with the TDIF peptide. The extracellular domain of TDR adopts a superhelical structure comprising 22 LRRs, and specifically recognizes TDIF by its inner concave surface. Together with our biochemical and sequence analyses, our structure reveals a conserved TDIF-recognition mechanism of TDR among plant species. Furthermore, a structural comparison of TDR with other plant LRR-RKs suggested the activation mechanism of TDR by TDIF. The structure of this CLE peptide receptor provides insights into the recognition mechanism of the CLE family peptides.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 2-11-16 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan.
Organizational Affiliation: