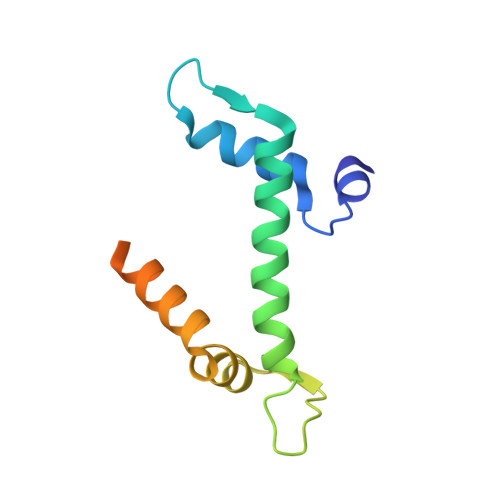
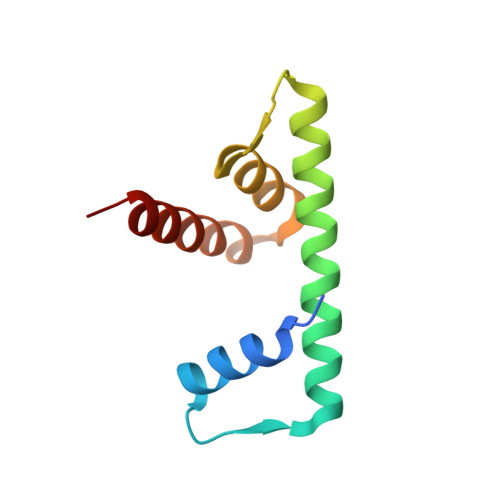
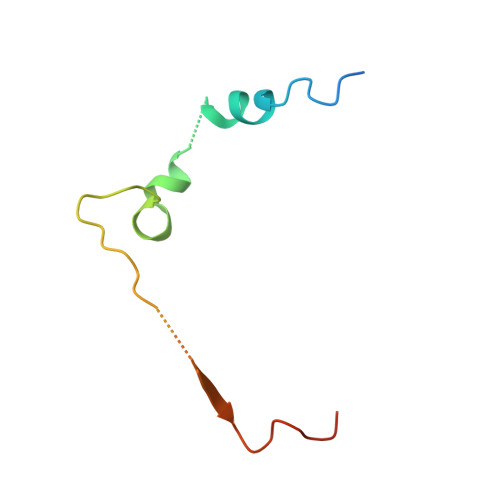
Molecular Basis and Specificity of H2A.Z-H2B Recognition and Deposition by the Histone Chaperone Yl1
Latrick, C.M., Marek, M., Ouararhni, K., Papin, C., Stoll, I., Ignatyeva, M., Obri, A., Ennifar, E., Dimitrov, S., Romier, C., Hamiche, A.(2016) Nat Struct Mol Biol 23: 309
- PubMed: 26974126
- DOI: https://doi.org/10.1038/nsmb.3189
- Primary Citation of Related Structures:
5FUG - PubMed Abstract:
H2A.Z, a widely conserved histone variant, is evicted from chromatin by the histone chaperone ANP32E. However, to date, no deposition chaperone for H2A.Z is known in metazoans. Here, we identify YL1 as a specific H2A.Z-deposition chaperone. The 2.7-Å-resolution crystal structure of the human YL1-H2A.Z-H2B complex shows that YL1 binding, similarly to ANP32E binding, triggers an extension of the H2A.Z αC helix. The interaction with YL1 is, however, more extensive and includes both the extended acidic patch and the entire DNA-binding surface of H2A.Z-H2B. Substitution of only four amino acid residues of H2A is sufficient for the formation of an H2A.Z-like interface specifically recognized by YL1. Collectively, our data reveal the molecular basis of H2A.Z-specific recognition by YL1 and shed light on the mechanism of H2A.Z transfer to the nucleosome by the ATP-dependent chromatin-remodeling complexes SRCAP and P400-TIP60.
- Equipe Labélisée Ligue Contre le Cancer, Département de Génomique Fonctionnelle et Cancer, Institut de Génétique et Biologie Moléculaire et Cellulaire (IGBMC), Illkirch, France.
Organizational Affiliation: