Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Jackson, V.A., Mehmood, S., Chavent, M., Roversi, P., Carrasquero, M., Del Toro, D., Seyit-Bremer, G., Ranaivoson, F.M., Comoletti, D., Sansom, M.S.P., Robinson, C.V., Klein, R., Seiradake, E.(2016) Nat Commun 7: 11184
- PubMed: 27091502
- DOI: https://doi.org/10.1038/ncomms11184
- Primary Citation of Related Structures:
5FTT, 5FTU - PubMed Abstract:
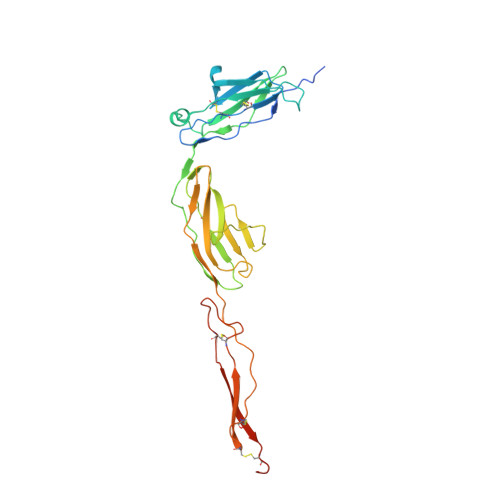
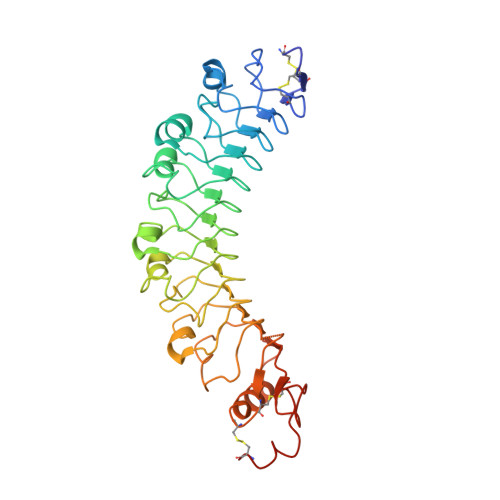
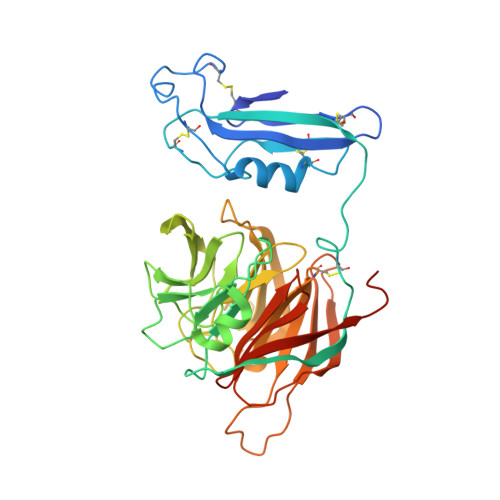
Latrophilin adhesion-GPCRs (Lphn1-3 or ADGRL1-3) and Unc5 cell guidance receptors (Unc5A-D) interact with FLRT proteins (FLRT1-3), thereby promoting cell adhesion and repulsion, respectively. How the three proteins interact and function simultaneously is poorly understood. We show that Unc5D interacts with FLRT2 in cis, controlling cell adhesion in response to externally presented Lphn3. The ectodomains of the three proteins bind cooperatively. Crystal structures of the ternary complex formed by the extracellular domains reveal that Lphn3 dimerizes when bound to FLRT2:Unc5, resulting in a stoichiometry of 1:1:2 (FLRT2:Unc5D:Lphn3). This 1:1:2 complex further dimerizes to form a larger 'super-complex' (2:2:4), using a previously undescribed binding motif in the Unc5D TSP1 domain. Molecular dynamics simulations, point-directed mutagenesis and mass spectrometry demonstrate the stability and molecular properties of these complexes. Our data exemplify how receptors increase their functional repertoire by forming different context-dependent higher-order complexes.
- Department of Biochemistry, Oxford University, Oxford OX1 3QU, UK.
Organizational Affiliation: