Data-Collection Strategy for Challenging Native Sad Phasing.
Olieric, V., Weinert, T., Finke, A.D., Anders, C., Li, D., Olieric, N., Borca, C.N., Steinmetz, M.O., Caffrey, M., Jinek, M., Wang, M.(2016) Acta Crystallogr D Biol Crystallogr 72: 421
- PubMed: 26960129
- DOI: https://doi.org/10.1107/S2059798315024110
- Primary Citation of Related Structures:
5DWK, 5FQ5 - PubMed Abstract:
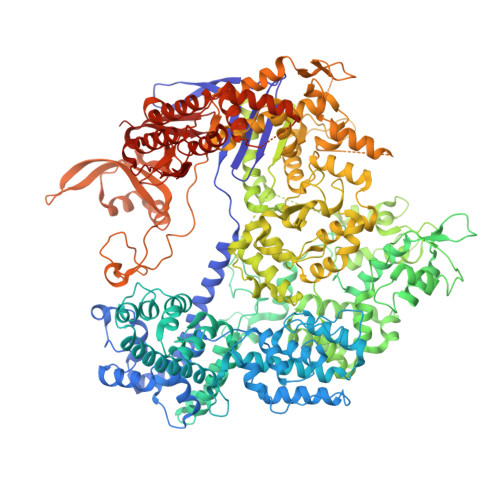
Recent improvements in data-collection strategies have pushed the limits of native SAD (single-wavelength anomalous diffraction) phasing, a method that uses the weak anomalous signal of light elements naturally present in macromolecules. These involve the merging of multiple data sets from either multiple crystals or from a single crystal collected in multiple orientations at a low X-ray dose. Both approaches yield data of high multiplicity while minimizing radiation damage and systematic error, thus ensuring accurate measurements of the anomalous differences. Here, the combined use of these two strategies is described to solve cases of native SAD phasing that were particular challenges: the integral membrane diacylglycerol kinase (DgkA) with a low Bijvoet ratio of 1% and the large 200 kDa complex of the CRISPR-associated endonuclease (Cas9) bound to guide RNA and target DNA crystallized in the low-symmetry space group C2. The optimal native SAD data-collection strategy based on systematic measurements performed on the 266 kDa multiprotein/multiligand tubulin complex is discussed.
- Swiss Light Source, Paul Scherrer Institut, Villigen PSI, Switzerland.
Organizational Affiliation: