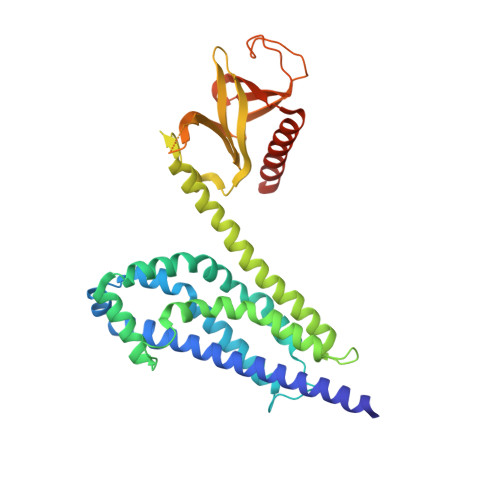
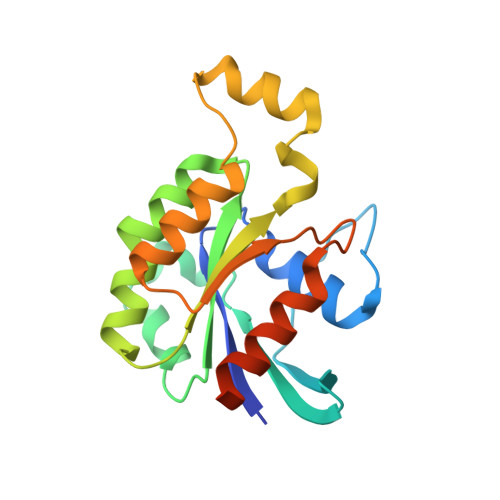
Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Cash, J.N., Davis, E.M., Tesmer, J.J.(2016) Structure 24: 730-740
- PubMed: 27150042
- DOI: https://doi.org/10.1016/j.str.2016.02.022
- Primary Citation of Related Structures:
5D27, 5D3V, 5D3W, 5D3X, 5D3Y, 5FI0, 5FI1 - PubMed Abstract:
Phosphatidylinositol 3,4,5-trisphosphate (PIP3)-dependent Rac exchanger 1 (P-Rex1) is a Rho guanine nucleotide exchange factor synergistically activated by PIP3 and Gβγ that plays an important role in the metastasis of breast, prostate, and skin cancer, making it an attractive therapeutic target. However, the molecular mechanisms behind P-Rex1 regulation are poorly understood. We determined structures of the P-Rex1 pleckstrin homology (PH) domain bound to the headgroup of PIP3 and resolved that PIP3 binding to the PH domain is required for P-Rex1 activity in cells but not for membrane localization, which points to an allosteric activation mechanism by PIP3. We also determined structures of the P-Rex1 tandem Dbl homology/PH domains in complexes with two of its substrate GTPases, Rac1 and Cdc42. Collectively, this study provides important molecular insights into P-Rex1 regulation and tools for targeting the PIP3-binding pocket of P-Rex1 with a new generation of cancer chemotherapeutic agents.
- Departments of Pharmacology and Biological Chemistry, Life Sciences Institute, University of Michigan, Ann Arbor, MI 48109-2216, USA.
Organizational Affiliation: