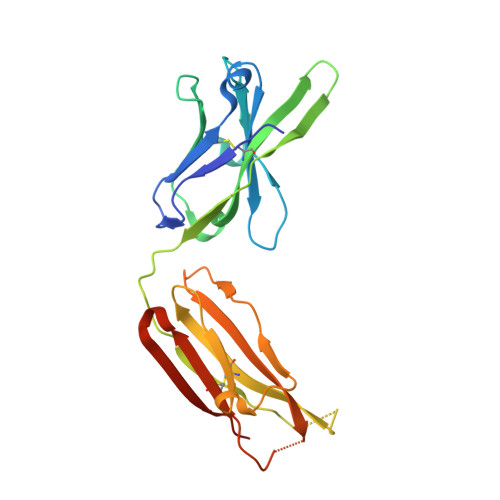
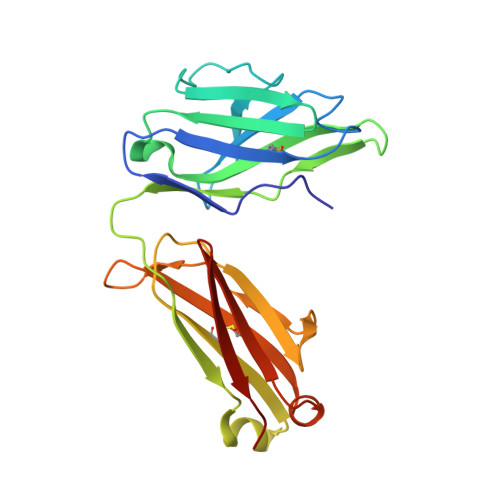
Paring Down HIV Env: Design and Crystal Structure of a Stabilized Inner Domain of HIV-1 gp120 Displaying a Major ADCC Target of the A32 Region.
Tolbert, W.D., Gohain, N., Veillette, M., Chapleau, J.P., Orlandi, C., Visciano, M.L., Ebadi, M., DeVico, A.L., Fouts, T.R., Finzi, A., Lewis, G.K., Pazgier, M.(2016) Structure 24: 697-709
- PubMed: 27041594
- DOI: https://doi.org/10.1016/j.str.2016.03.005
- Primary Citation of Related Structures:
4YBL, 4YC2, 5FCU - PubMed Abstract:
Evidence supports a role of antibody-dependent cellular cytotoxicity (ADCC) toward transitional epitopes in the first and second constant (C1-C2) regions of gp120 (A32-like epitopes) in preventing HIV-1 infection and in vaccine-induced protection. Here, we describe the first successful attempt at isolating the inner domain (ID) of gp120 as an independent molecule that encapsulates the A32-like region within a minimal structural unit of the HIV-1 Env. Through structure-based design, we developed ID2, which consists of the ID expressed independently of the outer domain and stabilized in the CD4-bound conformation by an inter-layer disulfide bond. ID2 expresses C1-C2 epitopes in the context of CD4-triggered full-length gp120 but without any known neutralizing epitope present. Thus, ID2 represents a novel probe for the analysis and/or selective induction of antibody responses to the A32 epitope region. We also present the crystal structure of ID2 complexed with mAb A32, which defines its epitope.
- Division of Vaccine Research, Institute of Human Virology, University of Maryland School of Medicine, Baltimore, MD 21201, USA; Department of Biochemistry and Molecular Biology, University of Maryland School of Medicine, Baltimore, MD 21201, USA.
Organizational Affiliation: