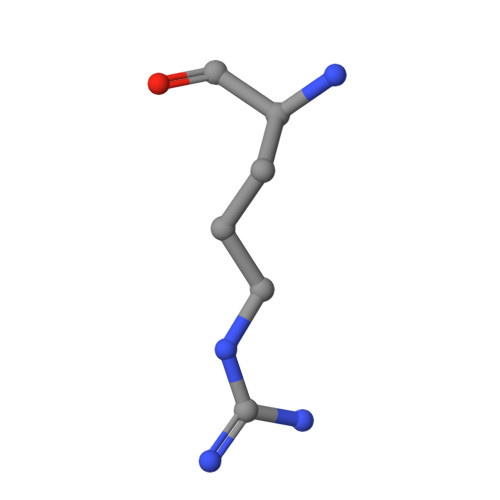
Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5.
Mavrakis, K.J., McDonald, E.R., Schlabach, M.R., Billy, E., Hoffman, G.R., deWeck, A., Ruddy, D.A., Venkatesan, K., Yu, J., McAllister, G., Stump, M., deBeaumont, R., Ho, S., Yue, Y., Liu, Y., Yan-Neale, Y., Yang, G., Lin, F., Yin, H., Gao, H., Kipp, D.R., Zhao, S., McNamara, J.T., Sprague, E.R., Zheng, B., Lin, Y., Cho, Y.S., Gu, J., Crawford, K., Ciccone, D., Vitari, A.C., Lai, A., Capka, V., Hurov, K., Porter, J.A., Tallarico, J., Mickanin, C., Lees, E., Pagliarini, R., Keen, N., Schmelzle, T., Hofmann, F., Stegmeier, F., Sellers, W.R.(2016) Science 351: 1208-1213
- PubMed: 26912361
- DOI: https://doi.org/10.1126/science.aad5944
- Primary Citation of Related Structures:
5FA5 - PubMed Abstract:
5-Methylthioadenosine phosphorylase (MTAP) is a key enzyme in the methionine salvage pathway. The MTAP gene is frequently deleted in human cancers because of its chromosomal proximity to the tumor suppressor gene CDKN2A. By interrogating data from a large-scale short hairpin RNA-mediated screen across 390 cancer cell line models, we found that the viability of MTAP-deficient cancer cells is impaired by depletion of the protein arginine methyltransferase PRMT5. MTAP-deleted cells accumulate the metabolite methylthioadenosine (MTA), which we found to inhibit PRMT5 methyltransferase activity. Deletion of MTAP in MTAP-proficient cells rendered them sensitive to PRMT5 depletion. Conversely, reconstitution of MTAP in an MTAP-deficient cell line rescued PRMT5 dependence. Thus, MTA accumulation in MTAP-deleted cancers creates a hypomorphic PRMT5 state that is selectively sensitized toward further PRMT5 inhibition. Inhibitors of PRMT5 that leverage this dysregulated metabolic state merit further investigation as a potential therapy for MTAP/CDKN2A-deleted tumors.
- Novartis Institutes for Biomedical Research, Cambridge, MA 02139, USA.
Organizational Affiliation: