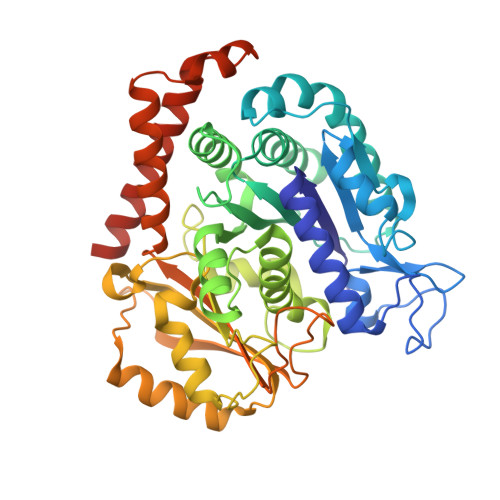
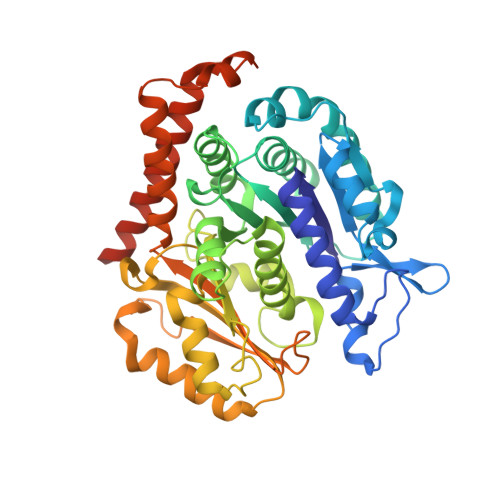
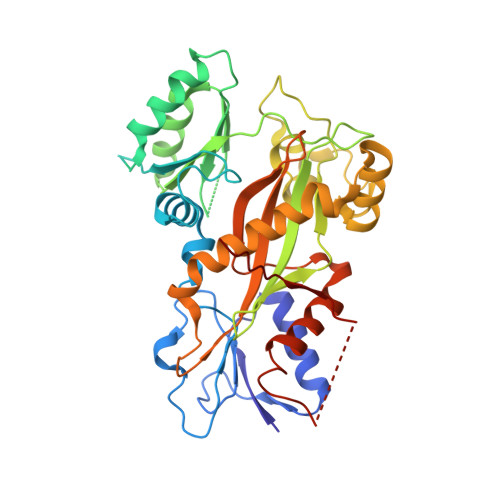
Mechanism of microtubule stabilization by taccalonolide AJ
Wang, Y., Yu, Y., Li, G.B., Li, S.A., Wu, C., Gigant, B., Qin, W., Chen, H., Wu, Y., Chen, Q., Yang, J.(2017) Nat Commun 8: 15787-15787
- PubMed: 28585532
- DOI: https://doi.org/10.1038/ncomms15787
- Primary Citation of Related Structures:
5EZY - PubMed Abstract:
As a major component of the cytoskeleton, microtubules consist of αβ-tubulin heterodimers and have been recognized as attractive targets for cancer chemotherapy. Microtubule-stabilizing agents (MSAs) promote polymerization of tubulin and stabilize the polymer, preventing depolymerization. The molecular mechanisms by which MSAs stabilize microtubules remain elusive. Here we report a 2.05 Å crystal structure of tubulin complexed with taccalonolide AJ, a newly identified taxane-site MSA. Taccalonolide AJ covalently binds to β-tubulin D226. On AJ binding, the M-loop undergoes a conformational shift to facilitate tubulin polymerization. In this tubulin-AJ complex, the E-site of tubulin is occupied by GTP rather than GDP. Biochemical analyses confirm that AJ inhibits the hydrolysis of the E-site GTP. Thus, we propose that the β-tubulin E-site is locked into a GTP-preferred status by AJ binding. Our results provide experimental evidence for the connection between MSA binding and tubulin nucleotide state, and will help design new MSAs to overcome taxane resistance.
- State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, China.
Organizational Affiliation: