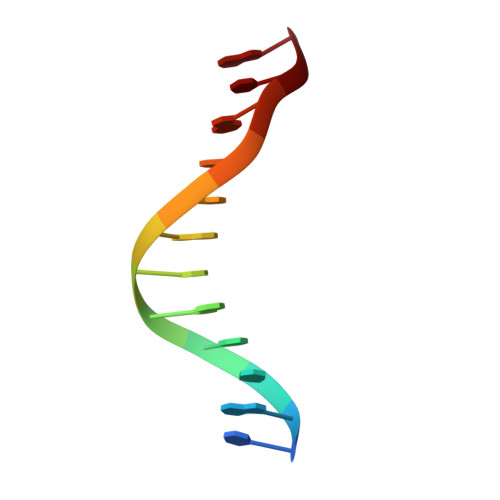
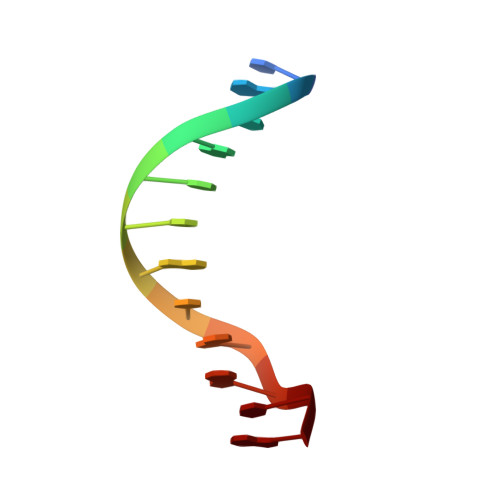
Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Mandal, P.K., Collie, G.W., Srivastava, S.C., Kauffmann, B., Huc, I.(2016) Nucleic Acids Res 44: 5936-5943
- PubMed: 27137886
- DOI: https://doi.org/10.1093/nar/gkw367
- Primary Citation of Related Structures:
5ET9, 5EWB, 5EYQ, 5EZF, 5F26, 5J0E - PubMed Abstract:
It has previously been shown that the use of racemic mixtures of naturally chiral macromolecules such as protein and DNA can significantly aid the crystallogenesis process, thereby addressing one of the major bottlenecks to structure determination by X-ray crystallographic methods-that of crystal growth. Although previous studies have provided convincing evidence of the applicability of the racemic crystallization technique to DNA through the study of well-characterized DNA structures, we sought to apply this method to a historically challenging DNA sequence. For this purpose we chose a non-self-complementary DNA duplex containing the biologically-relevant Pribnow box consensus sequence 'TATAAT'. Four racemic crystal structures of this previously un-crystallizable DNA target are reported (with resolutions in the range of 1.65-2.3 Å), with further crystallographic studies and structural analysis providing insight into the racemic crystallization process as well as structural details of this highly pertinent DNA sequence.
- Université de Bordeaux, CBMN (UMR5248), Pessac 33600, France CNRS, CBMN (UMR5248), Pessac 33600, France.
Organizational Affiliation: