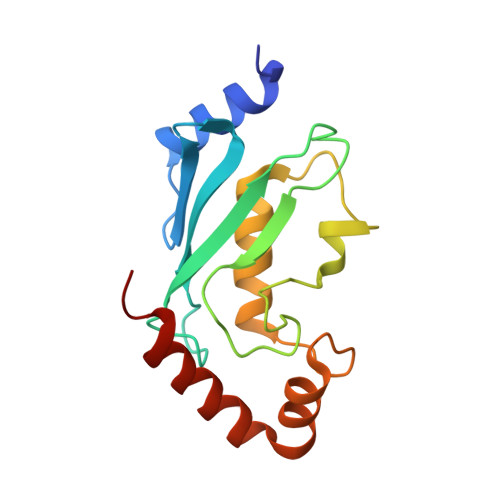
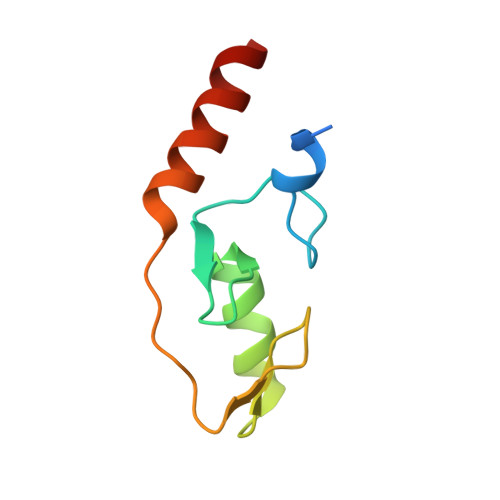
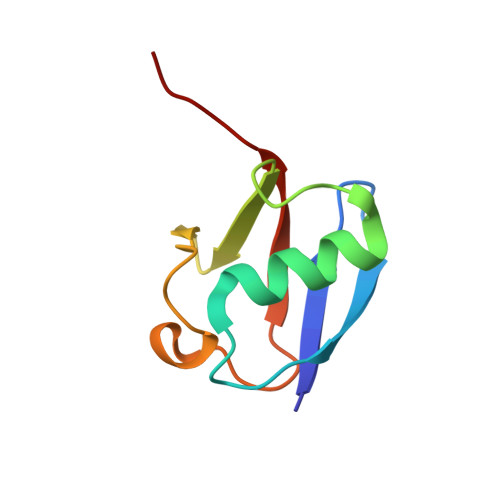
Mechanism of TRIM25 Catalytic Activation in the Antiviral RIG-I Pathway.
Sanchez, J.G., Chiang, J.J., Sparrer, K.M., Alam, S.L., Chi, M., Roganowicz, M.D., Sankaran, B., Gack, M.U., Pornillos, O.(2016) Cell Rep 16: 1315-1325
- PubMed: 27425606
- DOI: https://doi.org/10.1016/j.celrep.2016.06.070
- Primary Citation of Related Structures:
5EYA - PubMed Abstract:
Antiviral response pathways induce interferon by higher-order assembly of signaling complexes called signalosomes. Assembly of the RIG-I signalosome is regulated by K63-linked polyubiquitin chains, which are synthesized by the E3 ubiquitin ligase, TRIM25. We have previously shown that the TRIM25 coiled-coil domain is a stable, antiparallel dimer that positions two catalytic RING domains on opposite ends of an elongated rod. We now show that the RING domain is a separate self-association motif that engages ubiquitin-conjugated E2 enzymes as a dimer. RING dimerization is required for catalysis, TRIM25-mediated RIG-I ubiquitination, interferon induction, and antiviral activity. We also provide evidence that RING dimerization and E3 ligase activity are promoted by binding of the TRIM25 SPRY domain to the RIG-I effector domain. These results indicate that TRIM25 actively participates in higher-order assembly of the RIG-I signalosome and helps to fine-tune the efficiency of the RIG-I-mediated antiviral response.
- Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA 22908, USA.
Organizational Affiliation: