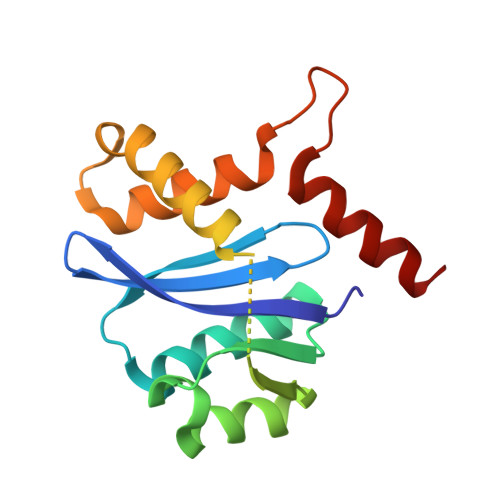
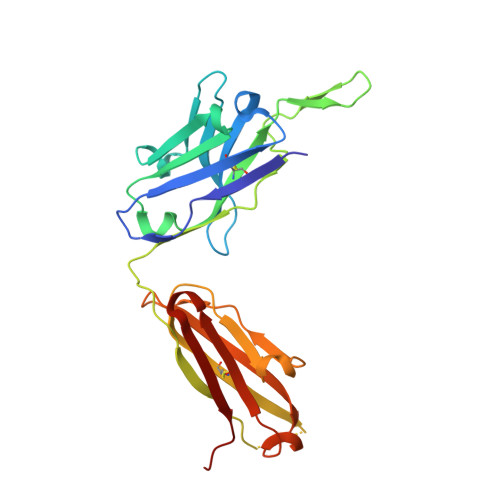
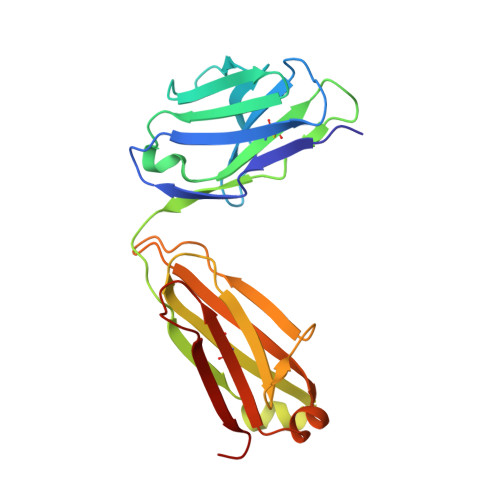
The Preserved HTH-Docking Cleft of HIV-1 Integrase Is Functionally Critical.
Galilee, M., Britan-Rosich, E., Griner, S.L., Uysal, S., Baumgartel, V., Lamb, D.C., Kossiakoff, A.A., Kotler, M., Stroud, R.M., Marx, A., Alian, A.(2016) Structure 24: 1936-1946
- PubMed: 27692964
- DOI: https://doi.org/10.1016/j.str.2016.08.015
- Primary Citation of Related Structures:
5EU7 - PubMed Abstract:
HIV-1 integrase (IN) catalyzes viral DNA integration into the host genome and facilitates multifunctional steps including virus particle maturation. Competency of IN to form multimeric assemblies is functionally critical, presenting an approach for anti-HIV strategies. Multimerization of IN depends on interactions between the distinct subunit domains and among the flanking protomers. Here, we elucidate an overlooked docking cleft of IN core domain that anchors the N-terminal helix-turn-helix (HTH) motif in a highly preserved and functionally critical configuration. Crystallographic structure of IN core domain in complex with Fab specifically targeting this cleft reveals a steric overlap that would inhibit HTH-docking, C-terminal domain contacts, DNA binding, and subsequent multimerization. While Fab inhibits in vitro IN integration activity, in vivo it abolishes virus particle production by specifically associating with preprocessed IN within Gag-Pol and interfering with early cytosolic Gag/Gag-Pol assemblies. The HTH-docking cleft may offer a fresh hotspot for future anti-HIV intervention strategies.
- Department of Biology, Technion - Israel Institute of Technology, Haifa 320003, Israel.
Organizational Affiliation: