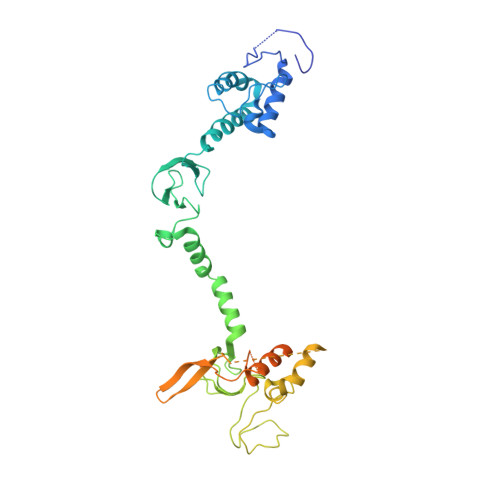
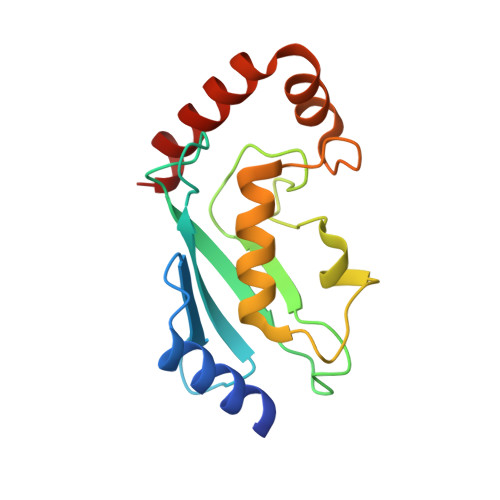
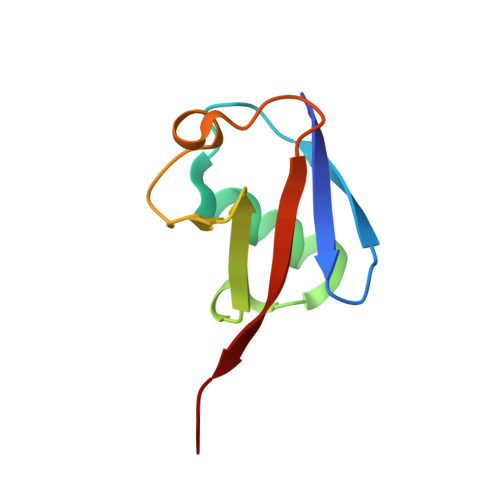
Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation.
Lechtenberg, B.C., Rajput, A., Sanishvili, R., Dobaczewska, M.K., Ware, C.F., Mace, P.D., Riedl, S.J.(2016) Nature 529: 546-550
- PubMed: 26789245
- DOI: https://doi.org/10.1038/nature16511
- Primary Citation of Related Structures:
5EDV - PubMed Abstract:
Ubiquitination is a central process affecting all facets of cellular signalling and function. A critical step in ubiquitination is the transfer of ubiquitin from an E2 ubiquitin-conjugating enzyme to a substrate or a growing ubiquitin chain, which is mediated by E3 ubiquitin ligases. RING-type E3 ligases typically facilitate the transfer of ubiquitin from the E2 directly to the substrate. The RING-between-RING (RBR) family of RING-type E3 ligases, however, breaks this paradigm by forming a covalent intermediate with ubiquitin similarly to HECT-type E3 ligases. The RBR family includes Parkin and HOIP, the central catalytic factor of the LUBAC (linear ubiquitin chain assembly complex). While structural insights into the RBR E3 ligases Parkin and HHARI in their overall auto-inhibited forms are available, no structures exist of intact fully active RBR E3 ligases or any of their complexes. Thus, the RBR mechanism of action has remained largely unknown. Here we present the first structure, to our knowledge, of the fully active human HOIP RBR in its transfer complex with an E2~ubiquitin conjugate, which elucidates the intricate nature of RBR E3 ligases. The active HOIP RBR adopts a conformation markedly different from that of auto-inhibited RBRs. HOIP RBR binds the E2~ubiquitin conjugate in an elongated fashion, with the E2 and E3 catalytic centres ideally aligned for ubiquitin transfer, which structurally both requires and enables a HECT-like mechanism. In addition, three distinct helix-IBR-fold motifs inherent to RBRs form ubiquitin-binding regions that engage the activated ubiquitin of the E2~ubiquitin conjugate and, surprisingly, an additional regulatory ubiquitin molecule. The features uncovered reveal critical states of the HOIP RBR E3 ligase cycle, and comparison with Parkin and HHARI suggests a general mechanism for RBR E3 ligases.
- NCI-Designated Cancer Center, Sanford Burnham Prebys Medical Discovery Institute, 10901 North Torrey Pines Road, La Jolla, California 92037, USA.
Organizational Affiliation: