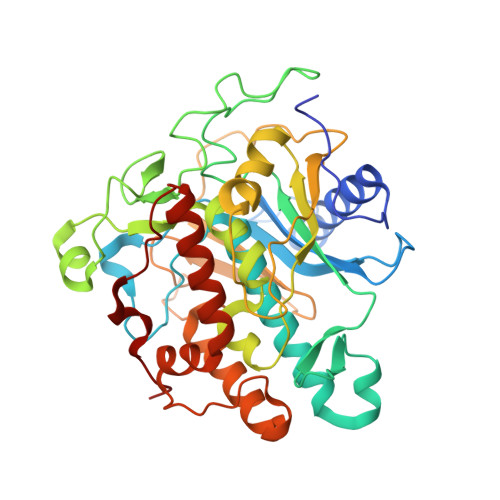
Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Matthews, M.M., Thomas, J.M., Zheng, Y., Tran, K., Phelps, K.J., Scott, A.I., Havel, J., Fisher, A.J., Beal, P.A.(2016) Nat Struct Mol Biol 23: 426-433
- PubMed: 27065196
- DOI: https://doi.org/10.1038/nsmb.3203
- Primary Citation of Related Structures:
5ED1, 5ED2, 5HP2, 5HP3 - PubMed Abstract:
Adenosine deaminases acting on RNA (ADARs) are editing enzymes that convert adenosine to inosine in duplex RNA, a modification reaction with wide-ranging consequences in RNA function. Understanding of the ADAR reaction mechanism, the origin of editing-site selectivity, and the effect of mutations is limited by the lack of high-resolution structural data for complexes of ADARs bound to substrate RNAs. Here we describe four crystal structures of the human ADAR2 deaminase domain bound to RNA duplexes bearing a mimic of the deamination reaction intermediate. These structures, together with structure-guided mutagenesis and RNA-modification experiments, explain the basis of the ADAR deaminase domain's dsRNA specificity, its base-flipping mechanism, and its nearest-neighbor preferences. In addition, we identified an ADAR2-specific RNA-binding loop near the enzyme active site, thus rationalizing differences in selectivity observed between different ADARs. Finally, our results provide a structural framework for understanding the effects of ADAR mutations associated with human disease.
- Department of Chemistry, University of California, Davis, Davis, California, USA.
Organizational Affiliation: