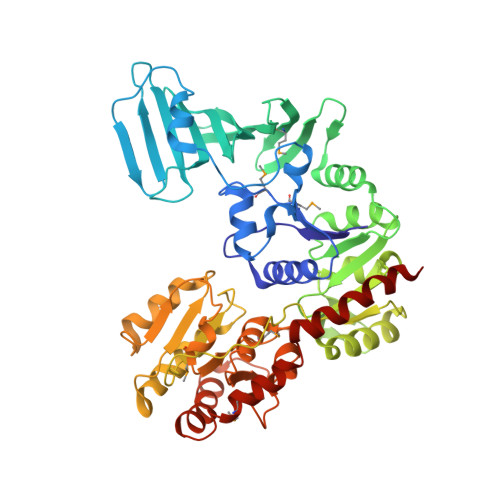
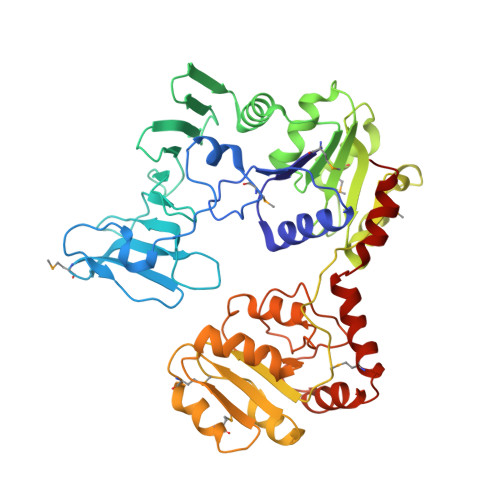
Mechanism of a cytosolic O-glycosyltransferase essential for the synthesis of a bacterial adhesion protein.
Chen, Y., Seepersaud, R., Bensing, B.A., Sullam, P.M., Rapoport, T.A.(2016) Proc Natl Acad Sci U S A 113: E1190-E1199
- PubMed: 26884191
- DOI: https://doi.org/10.1073/pnas.1600494113
- Primary Citation of Related Structures:
5E9T, 5E9U - PubMed Abstract:
O-glycosylation of Ser and Thr residues is an important process in all organisms, which is only poorly understood. Such modification is required for the export and function of adhesin proteins that mediate the attachment of pathogenic Gram-positive bacteria to host cells. Here, we have analyzed the mechanism by which the cytosolic O-glycosyltransferase GtfA/B of Streptococcus gordonii modifies the Ser/Thr-rich repeats of adhesin. The enzyme is a tetramer containing two molecules each of GtfA and GtfB. The two subunits have the same fold, but only GtfA contains an active site, whereas GtfB provides the primary binding site for adhesin. During a first phase of glycosylation, the conformation of GtfB is restrained by GtfA to bind substrate with unmodified Ser/Thr residues. In a slow second phase, GtfB recognizes residues that are already modified with N-acetylglucosamine, likely by converting into a relaxed conformation in which one interface with GtfA is broken. These results explain how the glycosyltransferase modifies a progressively changing substrate molecule.
- Howard Hughes Medical Institute, Harvard Medical School, Boston, MA 02115; Department of Cell Biology, Harvard Medical School, Boston, MA 02115;
Organizational Affiliation: