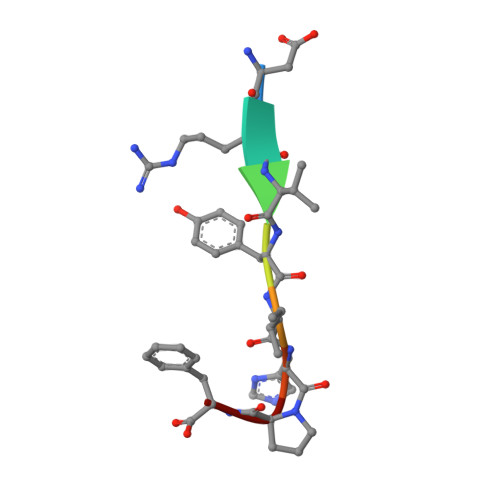
Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Kumar, P., Reithofer, V., Reisinger, M., Wallner, S., Pavkov-Keller, T., Macheroux, P., Gruber, K.(2016) Sci Rep 6: 23787-23787
- PubMed: 27025154
- DOI: https://doi.org/10.1038/srep23787
- Primary Citation of Related Structures:
5E2Q, 5E33, 5E3A, 5E3C, 5EGY, 5EHH - PubMed Abstract:
Human dipeptidyl-peptidase III (hDPP III) is a zinc-dependent hydrolase cleaving dipeptides off the N-termini of various bioactive peptides. Thus, the enzyme is likely involved in a number of physiological processes such as nociception and is also implicated in several forms of cancer. We present high-resolution crystal structures of hDPP III in complex with opioid peptides (Met-and Leu-enkephalin, endomorphin-2) as well as with angiotensin-II and the peptide inhibitor IVYPW. These structures confirm the previously reported large conformational change of the enzyme upon ligand binding and show that the structure of the closed conformation is independent of the nature of the bound peptide. The overall peptide-binding mode is also conserved ensuring the correct positioning of the scissile peptide bond with respect to the catalytic zinc ion. The structure of the angiotensin-II complex shows, how longer peptides are accommodated in the binding cleft of hDPP III. Differences in the binding modes allow a distinction between real substrates and inhibitory peptides or "slow" substrates. The latter displace a zinc bound water molecule necessitating the energetically much less favoured anhydride mechanism as opposed to the favoured promoted-water mechanism. The structural data also form the necessary framework for the design of specific hDPP III inhibitors.
- Institute of Molecular Biosciences, University of Graz, Humboldtstraße 50/3, 8010 Graz, Austria.
Organizational Affiliation: