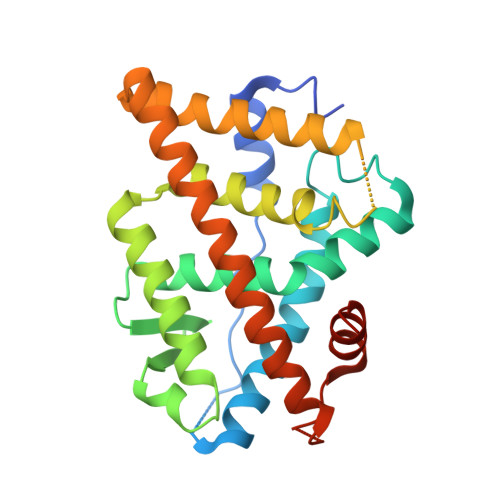
Predictive features of ligand-specific signaling through the estrogen receptor.
Nwachukwu, J.C., Srinivasan, S., Zheng, Y., Wang, S., Min, J., Dong, C., Liao, Z., Nowak, J., Wright, N.J., Houtman, R., Carlson, K.E., Josan, J.S., Elemento, O., Katzenellenbogen, J.A., Zhou, H.B., Nettles, K.W.(2016) Mol Syst Biol 12: 864-864
- PubMed: 27107013
- DOI: https://doi.org/10.15252/msb.20156701
- Primary Citation of Related Structures:
4ZN7, 4ZNH, 4ZNS, 4ZNT, 4ZNU, 4ZNV, 4ZNW, 5DI7, 5DID, 5DIE, 5DIG, 5DK9, 5DKB, 5DKE, 5DKG, 5DKS, 5DL4, 5DLR, 5DMC, 5DMF, 5DP0, 5DRJ, 5DRM, 5DTV, 5DU5, 5DUE, 5DUG, 5DUH, 5DVS, 5DVV, 5DWE, 5DWG, 5DWI, 5DWJ, 5DXK, 5DXM, 5DXP, 5DXQ, 5DXR, 5DY8, 5DYB, 5DYD, 5DZ0, 5DZ1, 5DZ3, 5DZH, 5DZI, 5E0W, 5E0X, 5E14 - PubMed Abstract:
Some estrogen receptor-α (ERα)-targeted breast cancer therapies such as tamoxifen have tissue-selective or cell-specific activities, while others have similar activities in different cell types. To identify biophysical determinants of cell-specific signaling and breast cancer cell proliferation, we synthesized 241 ERα ligands based on 19 chemical scaffolds, and compared ligand response using quantitative bioassays for canonical ERα activities and X-ray crystallography. Ligands that regulate the dynamics and stability of the coactivator-binding site in the C-terminal ligand-binding domain, called activation function-2 (AF-2), showed similar activity profiles in different cell types. Such ligands induced breast cancer cell proliferation in a manner that was predicted by the canonical recruitment of the coactivators NCOA1/2/3 and induction of the GREB1 proliferative gene. For some ligand series, a single inter-atomic distance in the ligand-binding domain predicted their proliferative effects. In contrast, the N-terminal coactivator-binding site, activation function-1 (AF-1), determined cell-specific signaling induced by ligands that used alternate mechanisms to control cell proliferation. Thus, incorporating systems structural analyses with quantitative chemical biology reveals how ligands can achieve distinct allosteric signaling outcomes through ERα.
- Department of Cancer Biology, The Scripps Research Institute, Jupiter, FL, USA.
Organizational Affiliation: