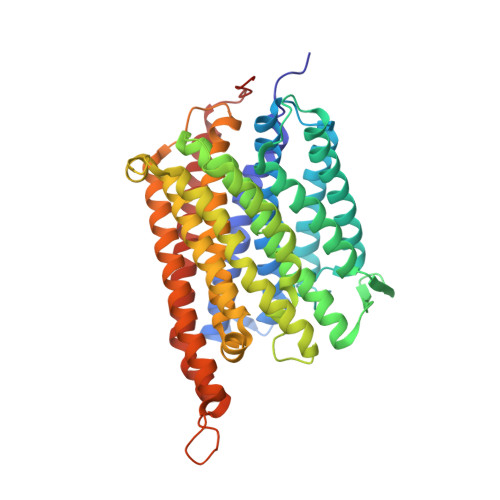
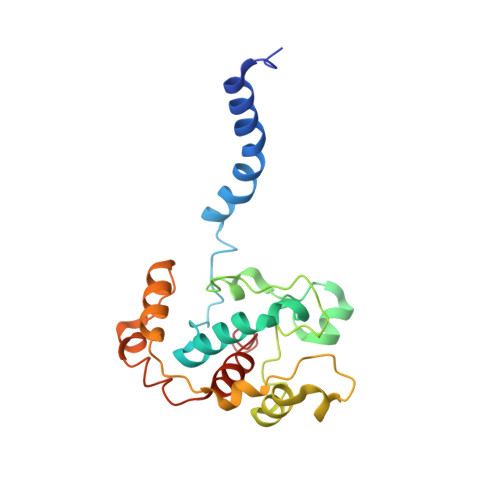
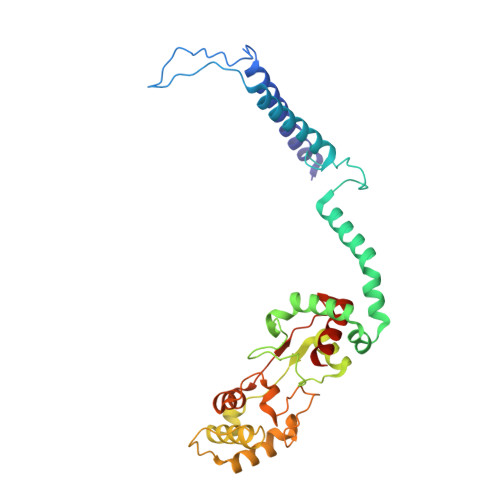
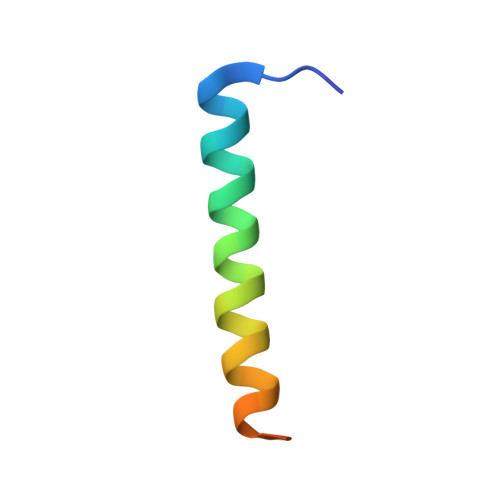
The structure of cbb3 cytochrome oxidase provides insights into proton pumping.
Buschmann, S., Warkentin, E., Xie, H., Langer, J.D., Ermler, U., Michel, H.(2010) Science 329: 327-330
- PubMed: 20576851
- DOI: https://doi.org/10.1126/science.1187303
- Primary Citation of Related Structures:
3MK7, 5DJQ - PubMed Abstract:
The heme-copper oxidases (HCOs) accomplish the key event of aerobic respiration; they couple O2 reduction and transmembrane proton pumping. To gain new insights into the still enigmatic process, we structurally characterized a C-family HCO--essential for the pathogenicity of many bacteria--that differs from the two other HCO families, A and B, that have been structurally analyzed. The x-ray structure of the C-family cbb3 oxidase from Pseudomonas stutzeri at 3.2 angstrom resolution shows an electron supply system different from families A and B. Like family-B HCOs, C HCOs have only one pathway, which conducts protons via an alternative tyrosine-histidine cross-link. Structural differences around hemes b and b3 suggest a different redox-driven proton-pumping mechanism and provide clues to explain the higher activity of family-C HCOs at low oxygen concentrations.
- Max-Planck-Institut für Biophysik, Max-von-Laue-Strasse 3, D-60438 Frankfurt/Main, Germany.
Organizational Affiliation: