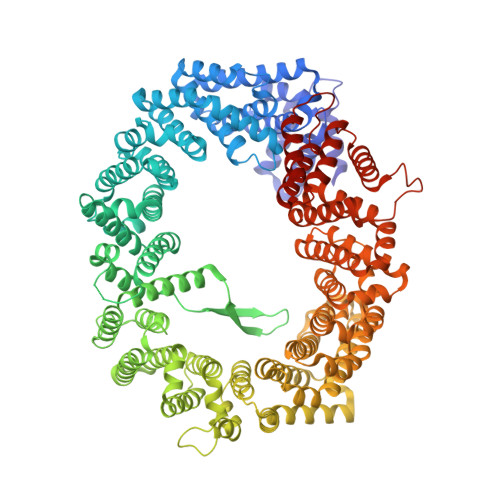
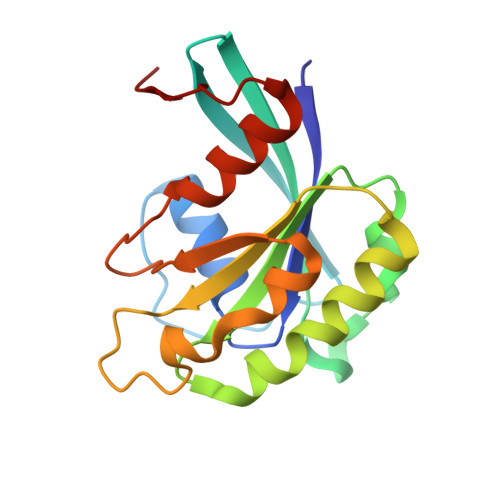
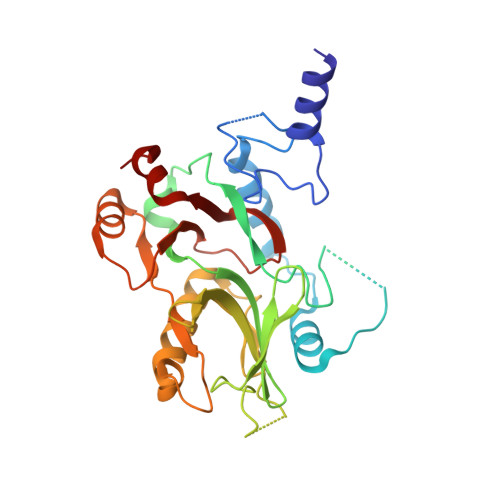
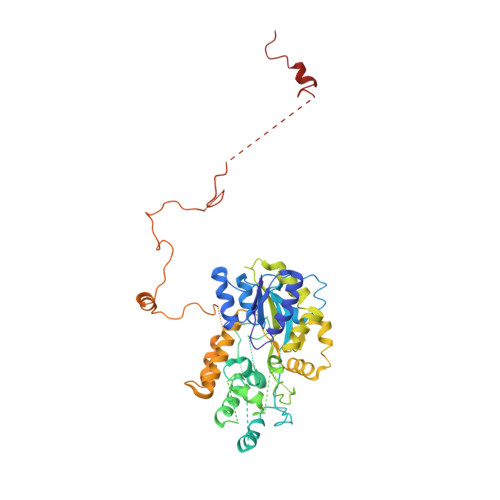
Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export.
Port, S.A., Monecke, T., Dickmanns, A., Spillner, C., Hofele, R., Urlaub, H., Ficner, R., Kehlenbach, R.H.(2015) Cell Rep 13: 690-702
- PubMed: 26489467
- DOI: https://doi.org/10.1016/j.celrep.2015.09.042
- Primary Citation of Related Structures:
5DIS - PubMed Abstract:
CRM1 is the major nuclear export receptor. During translocation through the nuclear pore, transport complexes transiently interact with phenylalanine-glycine (FG) repeats of multiple nucleoporins. On the cytoplasmic side of the nuclear pore, CRM1 tightly interacts with the nucleoporin Nup214. Here, we present the crystal structure of a 117-amino-acid FG-repeat-containing fragment of Nup214, in complex with CRM1, Snurportin 1, and RanGTP at 2.85 Å resolution. The structure reveals eight binding sites for Nup214 FG motifs on CRM1, with intervening stretches that are loosely attached to the transport receptor. Nup214 binds to N- and C-terminal regions of CRM1, thereby clamping CRM1 in a closed conformation and stabilizing the export complex. The role of conserved hydrophobic pockets for the recognition of FG motifs was analyzed in biochemical and cell-based assays. Comparative studies with RanBP3 and Nup62 shed light on specificities of CRM1-nucleoporin binding, which serves as a paradigm for transport receptor-nucleoporin interactions.
- Department of Molecular Biology, Faculty of Medicine, GZMB, Georg-August-University Göttingen, Humboldtallee 23, 37073 Göttingen, Germany.
Organizational Affiliation: