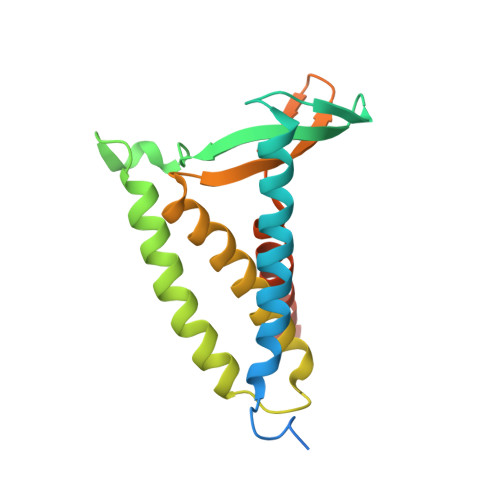
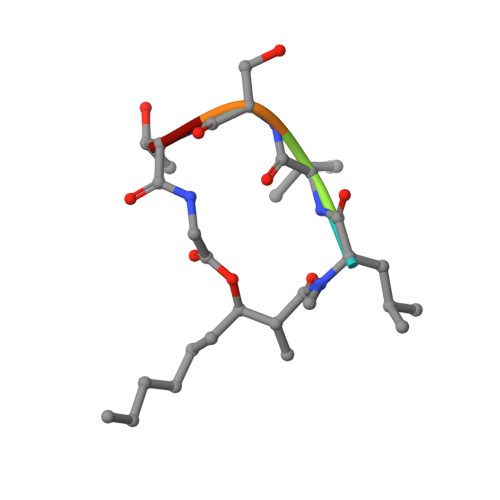
Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin.
Vogeley, L., El Arnaout, T., Bailey, J., Stansfeld, P.J., Boland, C., Caffrey, M.(2016) Science 351: 876-880
- PubMed: 26912896
- DOI: https://doi.org/10.1126/science.aad3747
- Primary Citation of Related Structures:
5DIR - PubMed Abstract:
With functions that range from cell envelope structure to signal transduction and transport, lipoproteins constitute 2 to 3% of bacterial genomes and play critical roles in bacterial physiology, pathogenicity, and antibiotic resistance. Lipoproteins are synthesized with a signal peptide securing them to the cytoplasmic membrane with the lipoprotein domain in the periplasm or outside the cell. Posttranslational processing requires a signal peptidase II (LspA) that removes the signal peptide. Here, we report the crystal structure of LspA from Pseudomonas aeruginosa complexed with the antimicrobial globomycin at 2.8 angstrom resolution. Mutagenesis studies identify LspA as an aspartyl peptidase. In an example of molecular mimicry, globomycin appears to inhibit by acting as a noncleavable peptide that sterically blocks the active site. This structure should inform rational antibiotic drug discovery.
- School of Medicine and School of Biochemistry and Immunology, Trinity College Dublin, Dublin, Ireland.
Organizational Affiliation: