Increased Conformational Flexibility of HLA-B*27 Subtypes Associated With Ankylosing Spondylitis.
Loll, B., Fabian, H., Huser, H., Hee, C.S., Ziegler, A., Uchanska-Ziegler, B., Ziegler, A.(2016) Arthritis Rheumatol 68: 1172-1182
- PubMed: 26748477
- DOI: https://doi.org/10.1002/art.39567
- Primary Citation of Related Structures:
5DEF, 5DEG - PubMed Abstract:
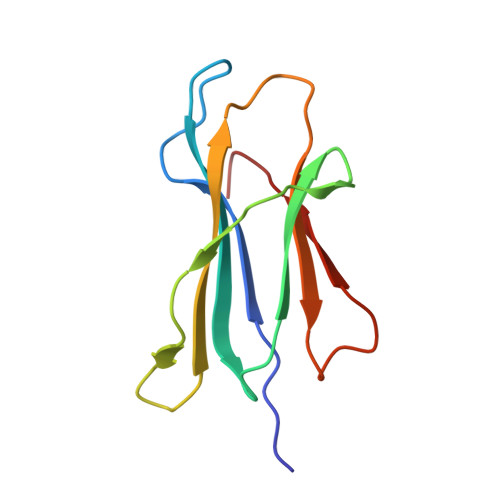
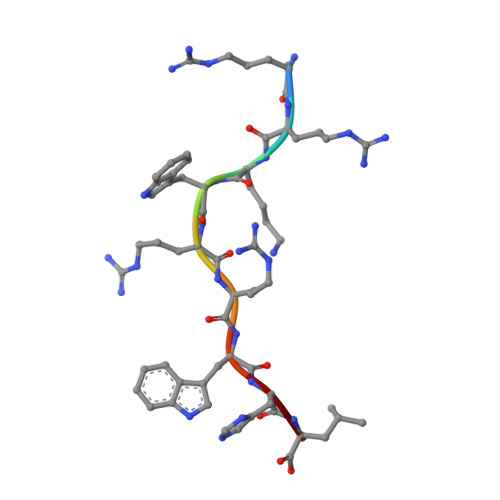
Dissimilarities in antigen processing and presentation are known to contribute to the differential association of HLA-B*27 subtypes with the inflammatory rheumatic disease ankylosing spondylitis (AS). In support of this notion, previous x-ray crystallographic data showed that peptides can be displayed by almost identical HLA-B*27 molecules in a subtype-dependent manner, allowing cytotoxic T lymphocytes to distinguish between these subtypes. For example, a human self-peptide derived from vasoactive intestinal peptide receptor type 1 (pVIPR; sequence RRKWRRWHL) is displayed in a single conformation by B*27:09 (which is not associated with AS), while B*27:05 (which is associated with AS) presents the peptide in a dual binding mode. In addition, differences in conformational flexibility between these subtypes might affect their stability or antigen presentation capability. This study was undertaken to investigate B*27:04 and B*27:06, another pair of minimally distinct HLA-B*27 subtypes, to assess whether dual peptide conformations or structural dynamics play a role in the initiation of AS. Using x-ray crystallography, we determined the structures of the pVIPR-B*27:04 and pVIPR-B*27:06 complexes and used isotope-edited infrared (IR) spectroscopy to probe the dynamics of these HLA-B*27 subtypes. As opposed to B*27:05 and B*27:09, B*27:04 (which is associated with AS) displays pVIPR conventionally and B*27:06 (which is not associated with AS) presents the peptide in a dual conformation. Comparison of the 4 HLA-B*27 subtypes using IR spectroscopy revealed that B*27:04 and B*27:05 possess elevated molecular dynamics compared to the nonassociated subtypes B*27:06 and B*27:09. Our results demonstrate that an increase in conformational flexibility characterizes the disease-associated subtypes B*27:04 and B*27:05.
- Freie Universität Berlin, Berlin, Germany.
Organizational Affiliation: