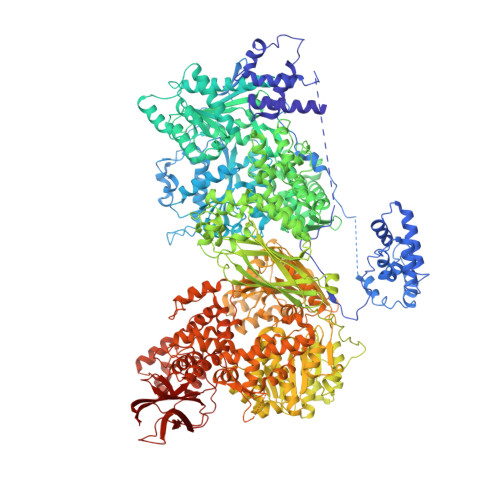
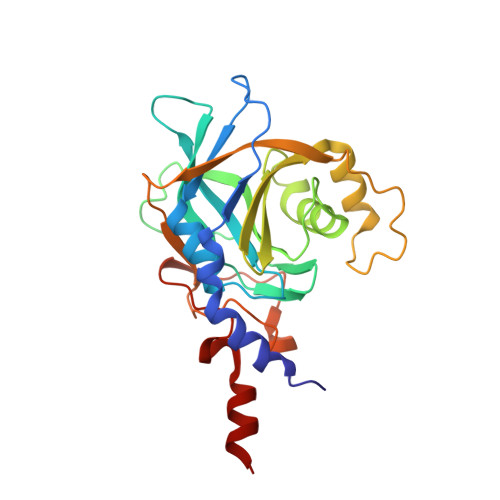
The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation.
Absmeier, E., Wollenhaupt, J., Mozaffari-Jovin, S., Becke, C., Lee, C.T., Preussner, M., Heyd, F., Urlaub, H., Luhrmann, R., Santos, K.F., Wahl, M.C.(2015) Genes Dev 29: 2576-2587
- PubMed: 26637280
- DOI: https://doi.org/10.1101/gad.271528.115
- Primary Citation of Related Structures:
5DCA - PubMed Abstract:
The Brr2 helicase provides the key remodeling activity for spliceosome catalytic activation, during which it disrupts the U4/U6 di-snRNP (small nuclear RNA protein), and its activity has to be tightly regulated. Brr2 exhibits an unusual architecture, including an ∼ 500-residue N-terminal region, whose functions and molecular mechanisms are presently unknown, followed by a tandem array of structurally similar helicase units (cassettes), only the first of which is catalytically active. Here, we show by crystal structure analysis of full-length Brr2 in complex with a regulatory Jab1/MPN domain of the Prp8 protein and by cross-linking/mass spectrometry of isolated Brr2 that the Brr2 N-terminal region encompasses two folded domains and adjacent linear elements that clamp and interconnect the helicase cassettes. Stepwise N-terminal truncations led to yeast growth and splicing defects, reduced Brr2 association with U4/U6•U5 tri-snRNPs, and increased ATP-dependent disruption of the tri-snRNP, yielding U4/U6 di-snRNP and U5 snRNP. Trends in the RNA-binding, ATPase, and helicase activities of the Brr2 truncation variants are fully rationalized by the crystal structure, demonstrating that the N-terminal region autoinhibits Brr2 via substrate competition and conformational clamping. Our results reveal molecular mechanisms that prevent premature and unproductive tri-snRNP disruption and suggest novel principles of Brr2-dependent splicing regulation.
- Laboratory of Structural Biochemistry, Freie Universität Berlin, D-14195 Berlin, Germany;
Organizational Affiliation: