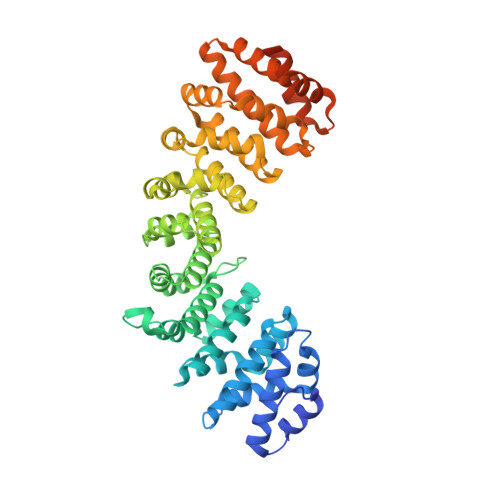
PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage.
Riccio, A.A., Cingolani, G., Pascal, J.M.(2016) Nucleic Acids Res 44: 1691-1702
- PubMed: 26704974
- DOI: https://doi.org/10.1093/nar/gkv1376
- Primary Citation of Related Structures:
5D5K - PubMed Abstract:
Poly(ADP-ribose) polymerase-2 (PARP-2) is one of three human PARP enzymes that are potently activated during the cellular DNA damage response (DDR). DDR-PARPs detect DNA strand breaks, leading to a dramatic increase in their catalytic production of the posttranslational modification poly(ADP-ribose) (PAR) to facilitate repair. There are limited biochemical and structural insights into the functional domains of PARP-2, which has restricted our understanding of how PARP-2 is specialized toward specific repair pathways. PARP-2 has a modular architecture composed of a C-terminal catalytic domain (CAT), a central Trp-Gly-Arg (WGR) domain and an N-terminal region (NTR). Although the NTR is generally considered the key DNA-binding domain of PARP-2, we report here that all three domains of PARP-2 collectively contribute to interaction with DNA damage. Biophysical, structural and biochemical analyses indicate that the NTR is natively disordered, and is only required for activation on specific types of DNA damage. Interestingly, the NTR is not essential for PARP-2 localization to sites of DNA damage. Rather, the WGR and CAT domains function together to recruit PARP-2 to sites of DNA breaks. Our study differentiates the functions of PARP-2 domains from those of PARP-1, the other major DDR-PARP, and highlights the specialization of the multi-domain architectures of DDR-PARPs.
- Department of Biochemistry and Molecular Biology, Sidney Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA 19107, USA.
Organizational Affiliation: