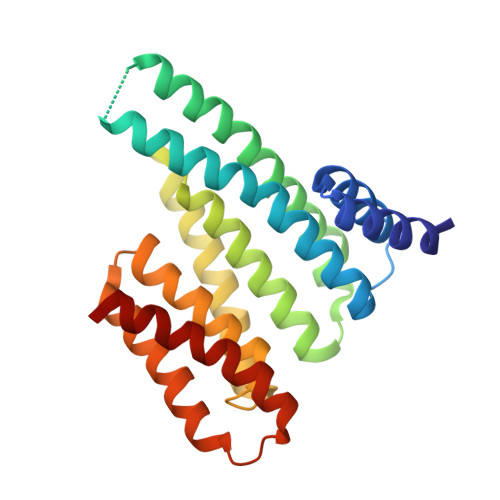
Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR.
Stevers, L.M., Lam, C.V., Leysen, S.F., Meijer, F.A., van Scheppingen, D.S., de Vries, R.M., Carlile, G.W., Milroy, L.G., Thomas, D.Y., Brunsveld, L., Ottmann, C.(2016) Proc Natl Acad Sci U S A 113: E1152-E1161
- PubMed: 26888287
- DOI: https://doi.org/10.1073/pnas.1516631113
- Primary Citation of Related Structures:
5D2D, 5D3E, 5D3F - PubMed Abstract:
Cystic fibrosis is a fatal genetic disease, most frequently caused by the retention of the CFTR (cystic fibrosis transmembrane conductance regulator) mutant protein in the endoplasmic reticulum (ER). The binding of the 14-3-3 protein to the CFTR regulatory (R) domain has been found to enhance CFTR trafficking to the plasma membrane. To define the mechanism of action of this protein-protein interaction, we have examined the interaction in vitro. The disordered multiphosphorylated R domain contains nine different 14-3-3 binding motifs. Furthermore, the 14-3-3 protein forms a dimer containing two amphipathic grooves that can potentially bind these phosphorylated motifs. This results in a number of possible binding mechanisms between these two proteins. Using multiple biochemical assays and crystal structures, we show that the interaction between them is governed by two binding sites: The key binding site of CFTR (pS768) occupies one groove of the 14-3-3 dimer, and a weaker, secondary binding site occupies the other binding groove. We show that fusicoccin-A, a natural-product tool compound used in studies of 14-3-3 biology, can stabilize the interaction between 14-3-3 and CFTR by selectively interacting with a secondary binding motif of CFTR (pS753). The stabilization of this interaction stimulates the trafficking of mutant CFTR to the plasma membrane. This definition of the druggability of the 14-3-3-CFTR interface might offer an approach for cystic fibrosis therapeutics.
- Laboratory of Chemical Biology, Department of Biomedical Engineering, Eindhoven University of Technology, 5600 MB Eindhoven, The Netherlands; Institute for Complex Molecular Systems, Eindhoven University of Technology, 5600 MB Eindhoven, The Netherlands;
Organizational Affiliation: