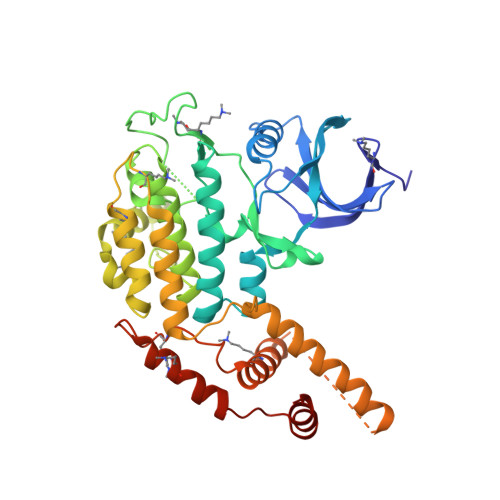
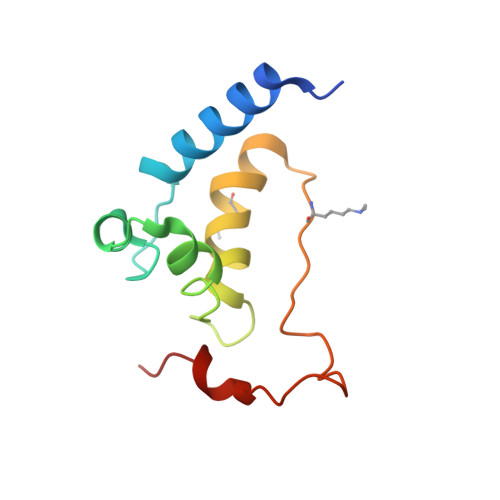
Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Ye, Q., Ur, S.N., Su, T.Y., Corbett, K.D.(2016) EMBO J 35: 2139-2151
- PubMed: 27491543
- DOI: https://doi.org/10.15252/embj.201694082
- Primary Citation of Related Structures:
4XH0, 4XHG, 4XHH, 4XHL, 5CYZ, 5CZO - PubMed Abstract:
In budding yeast, the monopolin complex mediates sister kinetochore cross-linking and co-orientation in meiosis I. The CK1δ kinase Hrr25 is critical for sister kinetochore co-orientation, but its roles are not well understood. Here, we present the structures of Hrr25 and its complex with the monopolin subunit Mam1. Hrr25 possesses a "central domain" that packs tightly against the kinase C-lobe, adjacent to the binding site for Mam1. Together, the Hrr25 central domain and Mam1 form a novel, contiguous embellishment to the Hrr25 kinase domain that affects Hrr25 conformational dynamics and enzyme kinetics. Mam1 binds a hydrophobic surface on the Hrr25 N-lobe that is conserved in CK1δ-family kinases, suggesting a role for this surface in recruitment and/or regulation of these enzymes throughout eukaryotes. Finally, using purified proteins, we find that Hrr25 phosphorylates the kinetochore receptor for monopolin, Dsn1. Together with our new structural insights into the fully assembled monopolin complex, this finding suggests that tightly localized Hrr25 activity modulates monopolin complex-kinetochore interactions through phosphorylation of both kinetochore and monopolin complex components.
- Ludwig Institute for Cancer Research, San Diego Branch, San Diego, La Jolla, CA, USA.
Organizational Affiliation: