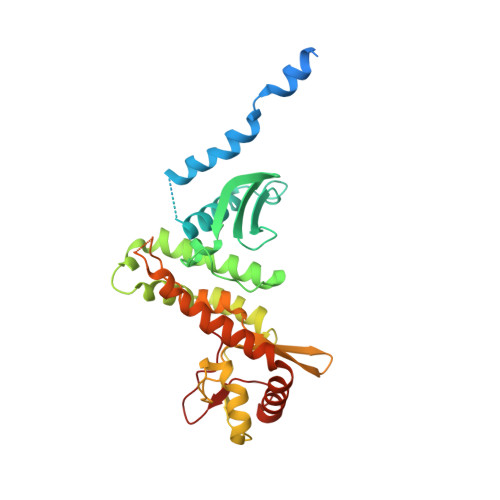
Salmonella Disrupts Host Endocytic Trafficking by SopD2-Mediated Inhibition of Rab7.
D'Costa, V.M., Braun, V., Landekic, M., Shi, R., Proteau, A., McDonald, L., Cygler, M., Grinstein, S., Brumell, J.H.(2015) Cell Rep 12: 1508-1518
- PubMed: 26299973
- DOI: https://doi.org/10.1016/j.celrep.2015.07.063
- Primary Citation of Related Structures:
5CPC, 5CQ9 - PubMed Abstract:
Intracellular bacterial pathogens of a diverse nature share the ability to evade host immunity by impairing trafficking of endocytic cargo to lysosomes for degradation, a process that is poorly understood. Here, we show that the Salmonella enterica type 3 secreted effector SopD2 mediates this process by binding the host regulatory GTPase Rab7 and inhibiting its nucleotide exchange. Consequently, this limits Rab7 interaction with its dynein- and kinesin-binding effectors RILP and FYCO1 and thereby disrupts host-driven regulation of microtubule motors. Our study identifies a bacterial effector capable of directly binding and thereby modulating Rab7 activity and a mechanism of endocytic trafficking disruption that may provide insight into the pathogenesis of other bacteria. Additionally, we provide a powerful tool for the study of Rab7 function, and a potential therapeutic target.
- Cell Biology Program, Hospital for Sick Children, Toronto, ON M5G 1X8, Canada.
Organizational Affiliation: