Locking the Elbow: Improved Antibody Fab Fragments as Chaperones for Structure Determination.
Bailey, L.J., Sheehy, K.M., Dominik, P.K., Liang, W.G., Rui, H., Clark, M., Jaskolowski, M., Kim, Y., Deneka, D., Tang, W.J., Kossiakoff, A.A.(2017) J Mol Biology
- PubMed: 29273204
- DOI: https://doi.org/10.1016/j.jmb.2017.12.012
- Primary Citation of Related Structures:
5CJO, 6AYZ, 6AZ2 - PubMed Abstract:
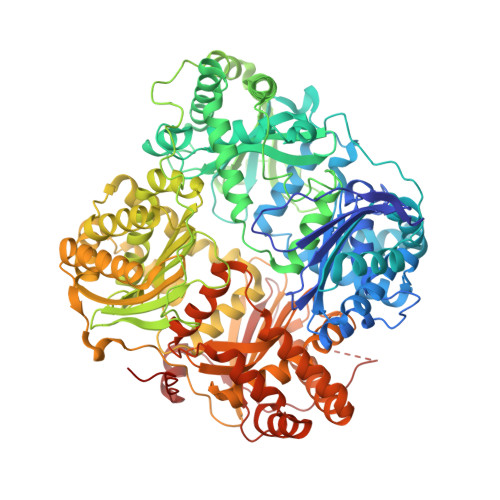
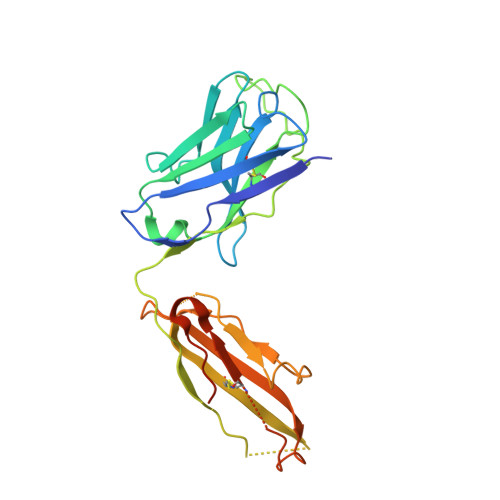
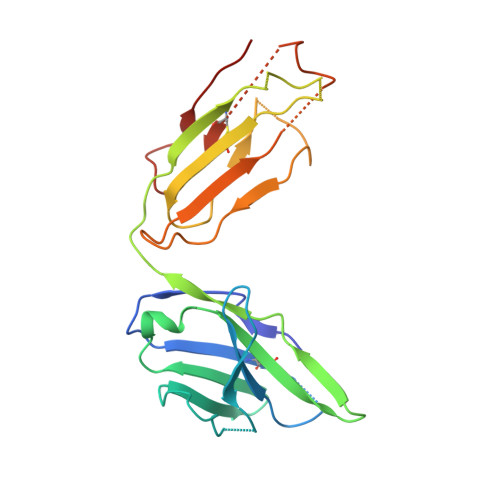
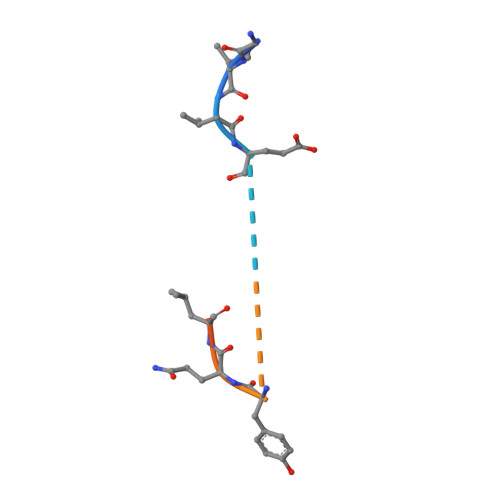
Antibody Fab fragments have been exploited with significant success to facilitate the structure determination of challenging macromolecules as crystallization chaperones and as molecular fiducial marks for single particle cryo-electron microscopy approaches. However, the inherent flexibility of the "elbow" regions, which link the constant and variable domains of the Fab, can introduce disorder and thus diminish their effectiveness. We have developed a phage display engineering strategy to generate synthetic Fab variants that significantly reduces elbow flexibility, while maintaining their high affinity and stability. This strategy was validated using previously recalcitrant Fab-antigen complexes where introduction of an engineered elbow region enhanced crystallization and diffraction resolution. Furthermore, incorporation of the mutations appears to be generally portable to other synthetic antibodies and may serve as a universal strategy to enhance the success rates of Fabs as structure determination chaperones.
- Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, IL 60637, USA.
Organizational Affiliation: