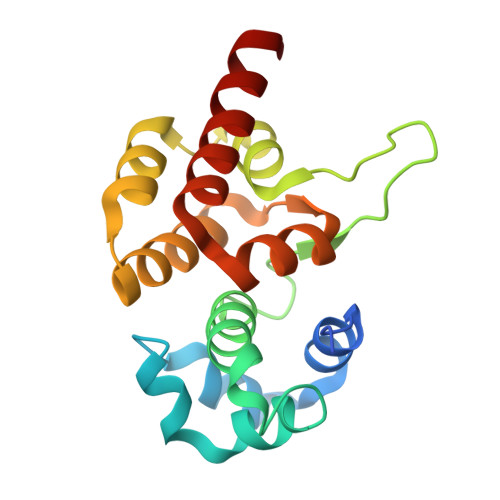
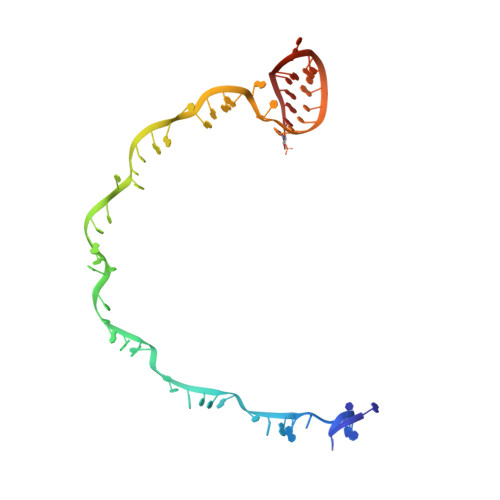
Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli.
van Erp, P.B., Jackson, R.N., Carter, J., Golden, S.M., Bailey, S., Wiedenheft, B.(2015) Nucleic Acids Res 43: 8381-8391
- PubMed: 26243775
- DOI: https://doi.org/10.1093/nar/gkv793
- Primary Citation of Related Structures:
5CD4 - PubMed Abstract:
In bacteria and archaea, short fragments of foreign DNA are integrated into Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) loci, providing a molecular memory of previous encounters with foreign genetic elements. In Escherichia coli, short CRISPR-derived RNAs are incorporated into a multi-subunit surveillance complex called Cascade (CRISPR-associated complex for antiviral defense). Recent structures of Cascade capture snapshots of this seahorse-shaped RNA-guided surveillance complex before and after binding to a DNA target. Here we determine a 3.2 Å x-ray crystal structure of Cascade in a new crystal form that provides insight into the mechanism of double-stranded DNA binding. Molecular dynamic simulations performed using available structures reveal functional roles for residues in the tail, backbone and belly subunits of Cascade that are critical for binding double-stranded DNA. Structural comparisons are used to make functional predictions and these predictions are tested in vivo and in vitro. Collectively, the results in this study reveal underlying mechanisms involved in target-induced conformational changes and highlight residues important in DNA binding and protospacer adjacent motif recognition.
- Department of Microbiology and Immunology, Montana State University, Bozeman, MT 59717, USA.
Organizational Affiliation: