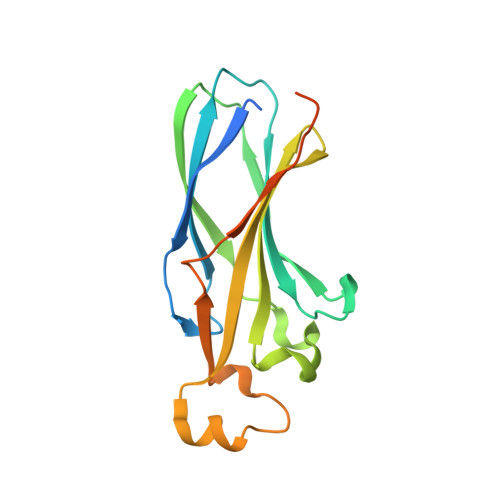
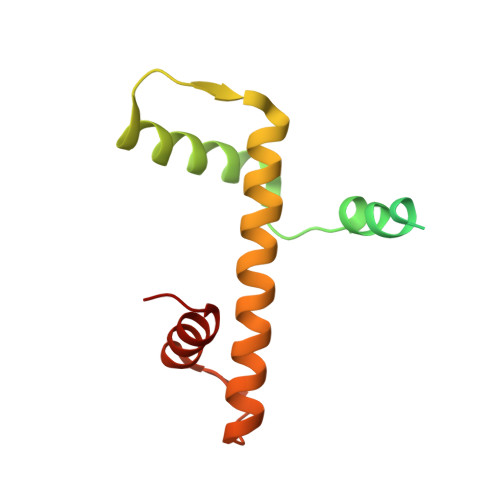
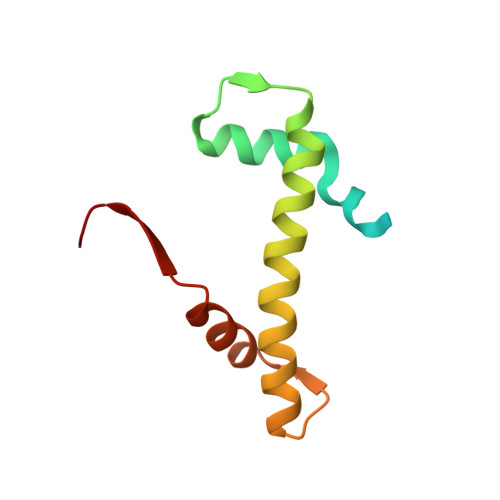
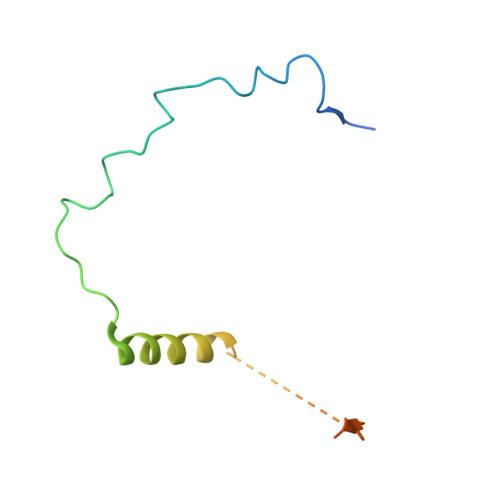
Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2
Wang, H., Wang, M., Yang, N., Xu, R.M.(2015) Protein Cell 6: 693-697
- PubMed: 26186914
- DOI: https://doi.org/10.1007/s13238-015-0190-0
- Primary Citation of Related Structures:
5C3I - National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China.
Organizational Affiliation: