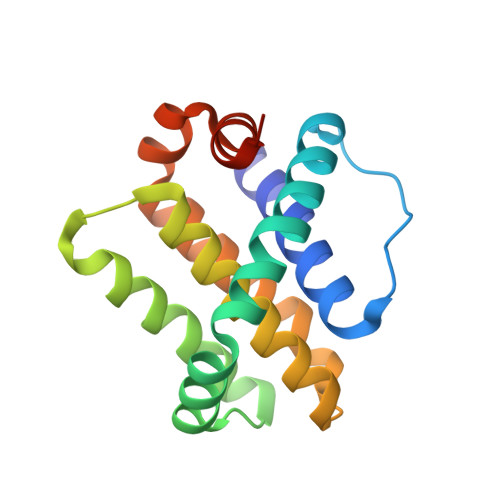
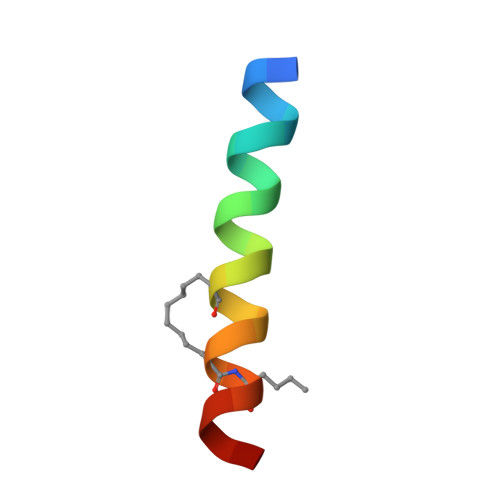
Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Miles, J.A., Yeo, D.J., Rowell, P., Rodriguez-Marin, S., Pask, C.M., Warriner, S.L., Edwards, T.A., Wilson, A.J.(2016) Chem Sci 7: 3694-3702
- PubMed: 28970875
- DOI: https://doi.org/10.1039/c5sc04048e
- Primary Citation of Related Structures:
5C3F, 5C3G - PubMed Abstract:
The development of constrained peptides represents an emerging strategy to generate peptide based probes and hits for drug-discovery that address challenging protein-protein interactions (PPIs). In this manuscript we report on the use of a novel α-alkenylglycine derived amino acid to synthesise hydrocarbon constrained BH3-family sequences (BIM and BID). Our biophysical and structural analyses illustrate that whilst the introduction of the constraint increases the population of the bioactive α-helical conformation of the peptide in solution, it does not enhance the inhibitory potency against pro-apoptotic Bcl-x L and Mcl-1 PPIs. SPR analyses indicate binding occurs via an induced fit mechanism whilst X-ray analyses illustrate none of the key interactions between the helix and protein are disturbed. The behaviour derives from enthalpy-entropy compensation which may be considered in terms of the ground state energies of the unbound constrained and unconstrained peptides; this has implications for the design of preorganised peptides to target protein-protein interactions.
- School of Chemistry , University of Leeds , Woodhouse Lane , Leeds LS2 9JT , UK . Email: A.J.Wilson@leeds.ac.uk.
Organizational Affiliation: