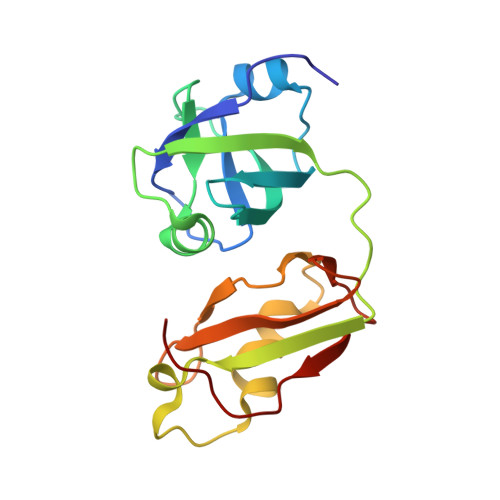
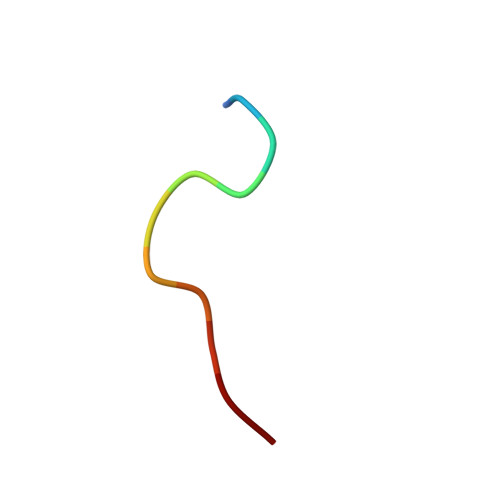Structural Details of Ufd1 Binding to p97 and Their Functional Implications in ER-Associated Degradation
Le, L.T.M., Kang, W., Kim, J.Y., Le, O.T.T., Lee, S.Y., Yang, J.K.(2016) PLoS One 11: e0163394-e0163394
- PubMed: 27684549
- DOI: https://doi.org/10.1371/journal.pone.0163394
- Primary Citation of Related Structures:
5B6C - PubMed Abstract:
The hexameric ATPase p97 has been implicated in diverse cellular processes through interactions with many different adaptor proteins at its N-terminal domain. Among these, the Ufd1-Npl4 heterodimer is a major adaptor, and the p97-Ufd1-Npl4 complex plays an essential role in endoplasmic reticulum-associated degradation (ERAD), acting as a segregase that translocates the ubiquitinated client protein from the ER membrane into the cytosol for proteasomal degradation. We determined the crystal structure of the complex of the N-terminal domain of p97 and the SHP box of Ufd1 at a resolution of 1.55 Å. The 11-residue-long SHP box of Ufd1 binds at the far-most side of the Nc lobe of the p97 N domain primarily through hydrophobic interactions, such that F225, F228, N233 and L235 of the SHP box contact hydrophobic residues on the surface of the p97 Nc lobe. Mutating these key interface residues abolished the interactions in two different binding experiments, isothermal titration calorimetry and co-immunoprecipitation. Furthermore, cycloheximide chase assays showed that these same mutations caused accumulation of tyrosinase-C89R, a well-known ERAD substrate, thus implying decreased rate of protein degradation due to their defects in ERAD function. Together, these results provide structural and biochemical insights into the interaction between p97 N domain and Ufd1 SHP box.
- Department of Chemistry, College of Natural Sciences, Soongsil University, Seoul 156-743, Korea.
Organizational Affiliation: