Monoubiquitination of histones H2B and H4 changes the nucleosome stability without affecting the nucleosome structure
Machida, S., Sekine, S., Nishiyama, Y., Horikoshi, N., Kurumizaka, H.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H3.2 | 139 | Homo sapiens | Mutation(s): 1 Gene Names: HIST2H3A, HIST2H3C, H3F2, H3FM, HIST2H3D | 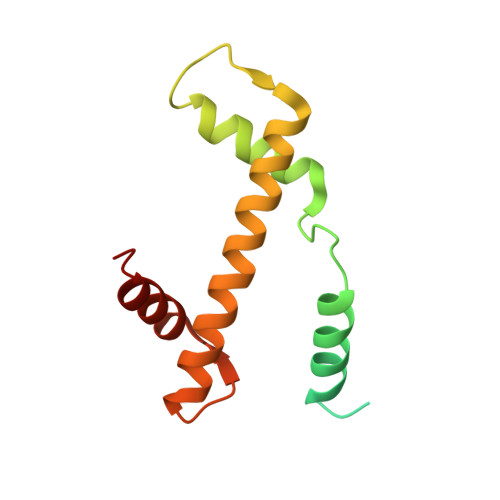 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q71DI3 (Homo sapiens) Explore Q71DI3 Go to UniProtKB: Q71DI3 | |||||
PHAROS: Q71DI3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q71DI3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H4 | 106 | Homo sapiens | Mutation(s): 1 Gene Names: | 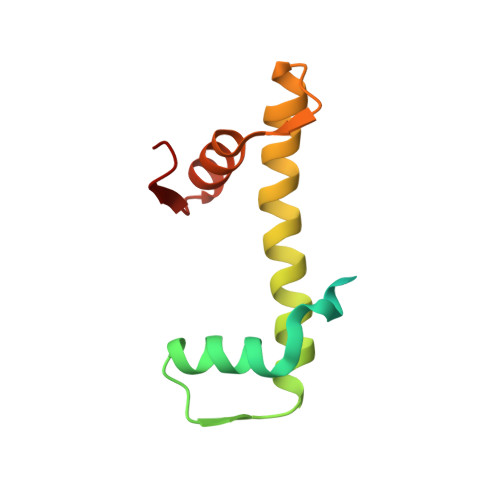 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62805 (Homo sapiens) Explore P62805 Go to UniProtKB: P62805 | |||||
PHAROS: P62805 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62805 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2A type 1-B/E | 133 | Homo sapiens | Mutation(s): 0 Gene Names: HIST1H2AB, H2AFM, HIST1H2AE, H2AFA | 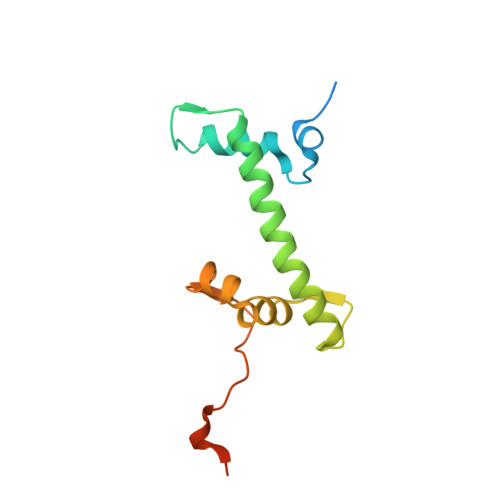 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P04908 (Homo sapiens) Explore P04908 Go to UniProtKB: P04908 | |||||
PHAROS: P04908 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04908 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2B type 1-J | 129 | Homo sapiens | Mutation(s): 1 Gene Names: HIST1H2BJ, H2BFR | 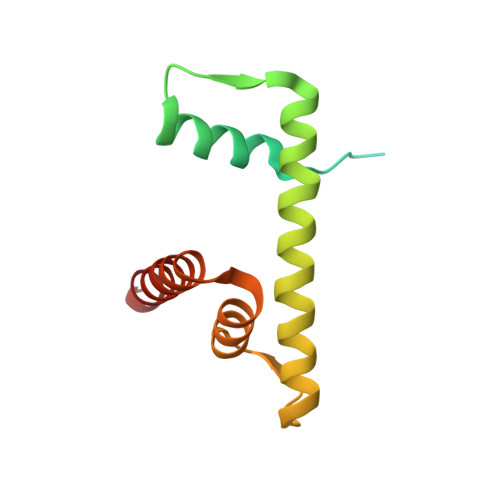 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P06899 (Homo sapiens) Explore P06899 Go to UniProtKB: P06899 | |||||
PHAROS: P06899 GTEx: ENSG00000124635 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06899 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (146-MER) | 146 | Homo sapiens |  | ||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 100.419 | α = 90 |
| b = 100.419 | β = 90 |
| c = 186.025 | γ = 120 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| MEXT KAKENHI | Japan | 25116002 |
| JSPS KAKENHI | Japan | 25250023 |
| JSPS KAKENHI | Japan | 26890023 |