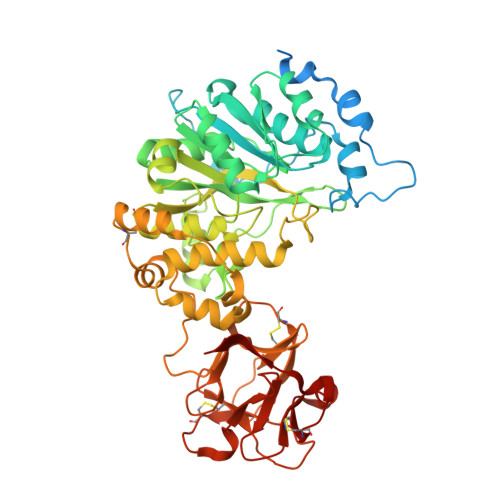
Dynamic Interplay between Catalytic and Lectin Domains of Galnac-Transferases Modulates Protein O-Glycosylation.
Lira-Navarrete, E., De Las Rivas, M., Companon, I., Pallares, M.C., Kong, Y., Iglesias-Fernandez, J., Bernardes, G.J.L., Peregrina, J.M., Rovira, C., Bernado, P., Bruscolini, P., Clausen, H., Lostao, A., Corzana, F., Hurtado-Guerrero, R.(2015) Nat Commun 6: 6937
- PubMed: 25939779
- DOI: https://doi.org/10.1038/ncomms7937
- Primary Citation of Related Structures:
5AJN, 5AJO, 5AJP - PubMed Abstract:
Protein O-glycosylation is controlled by polypeptide GalNAc-transferases (GalNAc-Ts) that uniquely feature both a catalytic and lectin domain. The underlying molecular basis of how the lectin domains of GalNAc-Ts contribute to glycopeptide specificity and catalysis remains unclear. Here we present the first crystal structures of complexes of GalNAc-T2 with glycopeptides that together with enhanced sampling molecular dynamics simulations demonstrate a cooperative mechanism by which the lectin domain enables free acceptor sites binding of glycopeptides into the catalytic domain. Atomic force microscopy and small-angle X-ray scattering experiments further reveal a dynamic conformational landscape of GalNAc-T2 and a prominent role of compact structures that are both required for efficient catalysis. Our model indicates that the activity profile of GalNAc-T2 is dictated by conformational heterogeneity and relies on a flexible linker located between the catalytic and the lectin domains. Our results also shed light on how GalNAc-Ts generate dense decoration of proteins with O-glycans.
- BIFI, University of Zaragoza, BIFI-IQFR (CSIC) Joint Unit, Mariano Esquillor s/n, Campus Rio Ebro, Edificio I+D, Zaragoza 50018, Spain.
Organizational Affiliation: