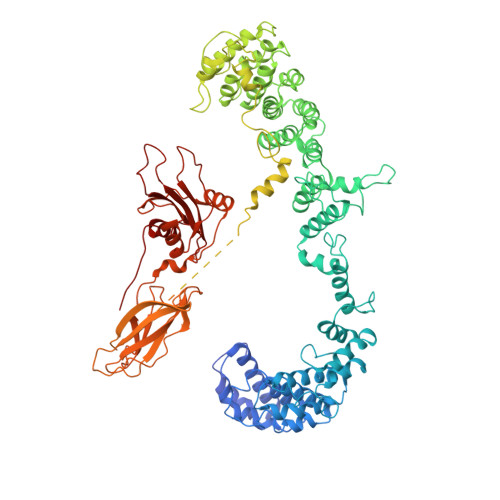
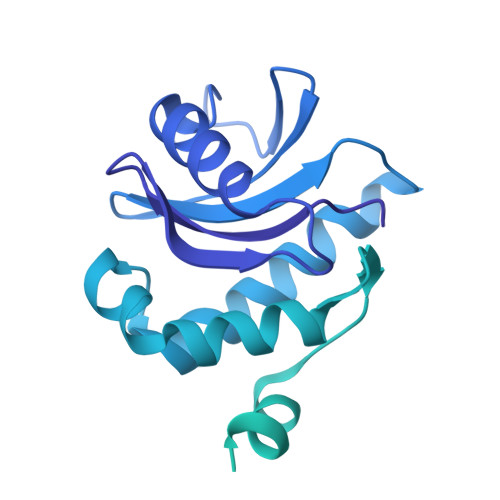
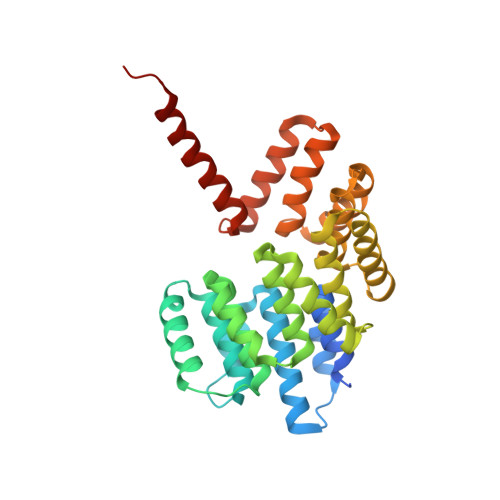
Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Dodonova, S.O., Diestelkoetter-Bachert, P., Von Appen, A., Hagen, W.J.H., Beck, R., Beck, M., Wieland, F., Briggs, J.A.G.(2015) Science 349: 195
- PubMed: 26160949
- DOI: https://doi.org/10.1126/science.aab1121
- Primary Citation of Related Structures:
5A1U, 5A1V, 5A1W, 5A1X, 5A1Y - PubMed Abstract:
Transport of material within cells is mediated by trafficking vesicles that bud from one cellular compartment and fuse with another. Formation of a trafficking vesicle is driven by membrane coats that localize cargo and polymerize into cages to bend the membrane. Although extensive structural information is available for components of these coats, the heterogeneity of trafficking vesicles has prevented an understanding of how complete membrane coats assemble on the membrane. We combined cryo-electron tomography, subtomogram averaging, and cross-linking mass spectrometry to derive a complete model of the assembled coat protein complex I (COPI) coat involved in traffic between the Golgi and the endoplasmic reticulum. The highly interconnected COPI coat structure contradicted the current "adaptor-and-cage" understanding of coated vesicle formation.
- Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Meyerhofstrasse 1, 69117 Heidelberg, Germany.
Organizational Affiliation: