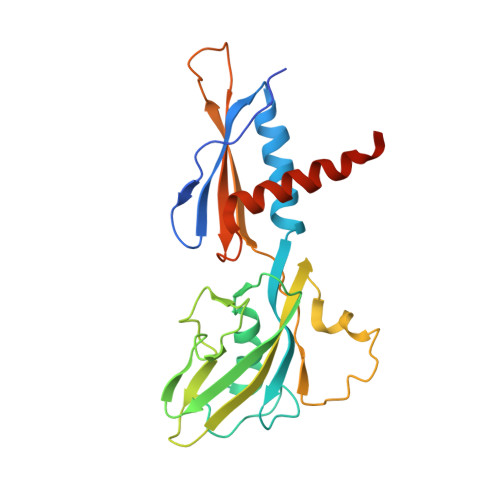
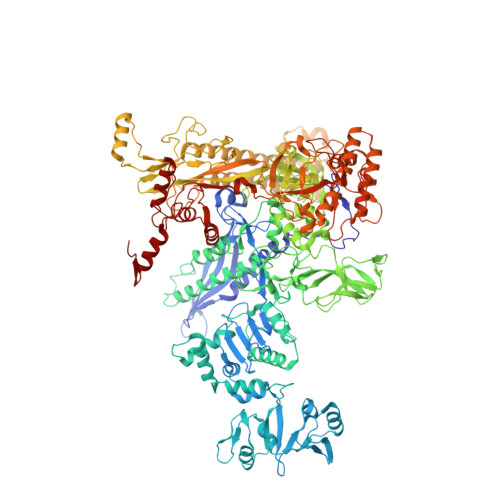
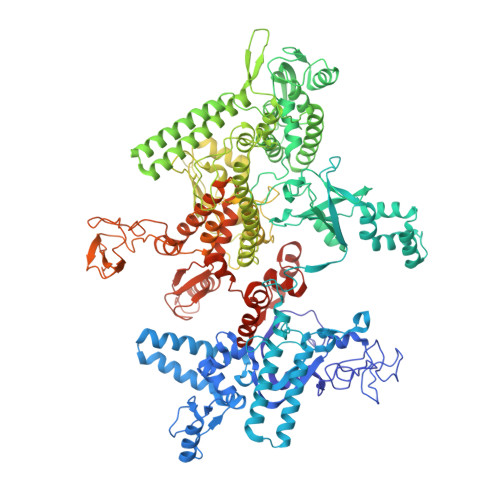
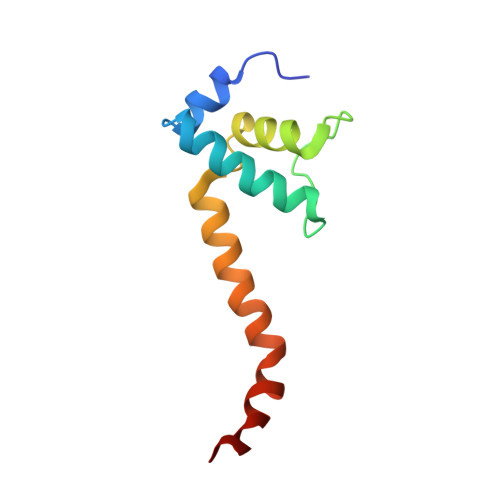
CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Bae, B., Nayak, D., Ray, A., Mustaev, A., Landick, R., Darst, S.A.(2015) Proc Natl Acad Sci U S A 112: E4178-E4187
- PubMed: 26195788
- DOI: https://doi.org/10.1073/pnas.1502368112
- Primary Citation of Related Structures:
4XSX, 4XSY, 4XSZ - PubMed Abstract:
RNA polymerase inhibitors like the CBR class that target the enzyme's complex catalytic center are attractive leads for new antimicrobials. Catalysis by RNA polymerase involves multiple rearrangements of bridge helix, trigger loop, and active-center side chains that isomerize the triphosphate of bound NTP and two Mg(2+) ions from a preinsertion state to a reactive configuration. CBR inhibitors target a crevice between the N-terminal portion of the bridge helix and a surrounding cap region within which the bridge helix is thought to rearrange during the nucleotide addition cycle. We report crystal structures of CBR inhibitor/Escherichia coli RNA polymerase complexes as well as biochemical tests that establish two distinct effects of the inhibitors on the RNA polymerase catalytic site. One effect involves inhibition of trigger-loop folding via the F loop in the cap, which affects both nucleotide addition and hydrolysis of 3'-terminal dinucleotides in certain backtracked complexes. The second effect is trigger-loop independent, affects only nucleotide addition and pyrophosphorolysis, and may involve inhibition of bridge-helix movements that facilitate reactive triphosphate alignment.
- The Rockefeller University, New York, NY 10065;
Organizational Affiliation: