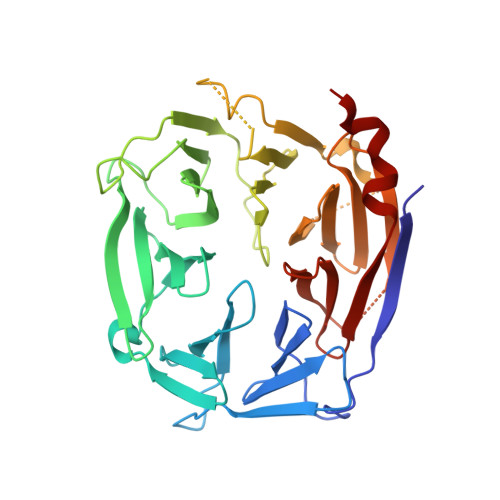
Structural Identification of the Vps18 Beta-Propeller Reveals a Critical Role in the Hops Complex Stability and Function.
Behrmann, H., Lurick, A., Kuhlee, A., Balderhaar, H.K., Brocker, C., Kummel, D., Engelbrecht-Vandre, S., Gohlke, U., Raunser, S., Heinemann, U., Ungermann, C.(2014) J Biological Chem 289: 33503
- PubMed: 25324549
- DOI: https://doi.org/10.1074/jbc.M114.602714
- Primary Citation of Related Structures:
4UUY - PubMed Abstract:
Membrane fusion at the vacuole, the lysosome equivalent in yeast, requires the HOPS tethering complex, which is recruited by the Rab7 GTPase Ypt7. HOPS provides a template for the assembly of SNAREs and thus likely confers fusion at a distinct position on vacuoles. Five of the six subunits in HOPS have a similar domain prediction with strong similarity to COPII subunits and nuclear porins. Here, we show that Vps18 indeed has a seven-bladed β-propeller as its N-terminal domain by revealing its structure at 2.14 Å. The Vps18 N-terminal domain can interact with the N-terminal part of Vps11 and also binds to lipids. Although deletion of the Vps18 N-terminal domain does not preclude HOPS assembly, as revealed by negative stain electron microscopy, the complex is instable and cannot support membrane fusion in vitro. We thus conclude that the β-propeller of Vps18 is required for HOPS stability and function and that it can serve as a starting point for further structural analyses of the HOPS tethering complex.
- From the Max-Delbrück Center for Molecular Medicine, Macromolecular Structure and Interaction Group, Robert-Rössle-Strasse 10, 13125 Berlin, Germany, Freie Universität Berlin, Chemistry and Biochemistry Institute, Takustrasse 6, 14195 Berlin, Germany.
Organizational Affiliation: