Boosting Affinity by Correct Ligand Preorganization for the S2 Pocket of Thrombin: A Study by Isothermal Titration Calorimetry, Molecular Dynamics, and High-Resolution Crystal Structures.
Ruhmann, E.H., Rupp, M., Betz, M., Heine, A., Klebe, G.(2016) ChemMedChem 11: 309
- PubMed: 26762840
- DOI: https://doi.org/10.1002/cmdc.201500531
- Primary Citation of Related Structures:
4UFD, 4UFE, 4UFF, 4UFG - PubMed Abstract:
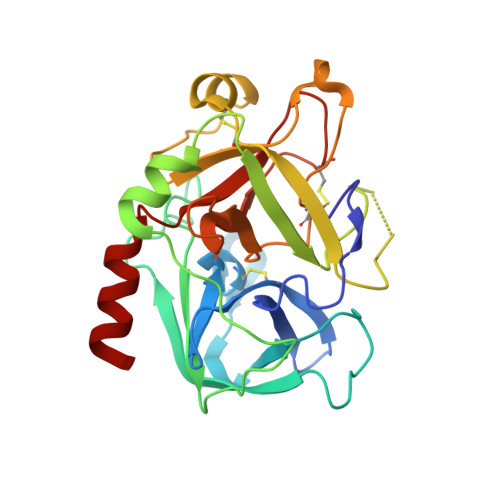
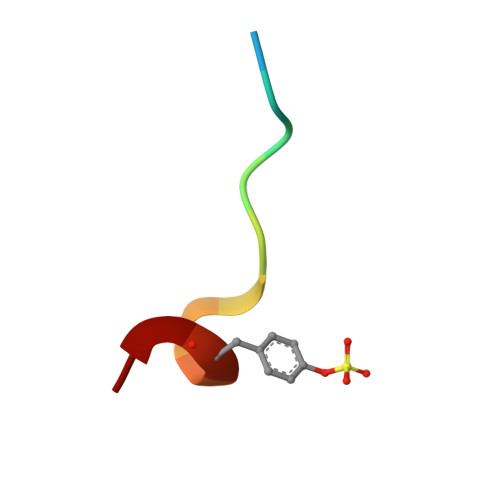
Structural preorganization to fix bioactive conformations at protein binding sites is a popular strategy to enhance binding affinity during late-stage optimization. The rationale for this enhancement relates to entropic advantages assigned to rigidified versus flexible ligands. We analyzed a narrow series of peptidomimetics binding to thrombin. The individual ligands exhibit at P2 a conformationally flexible glycine, more restricted alanine, N-methylglycine, N-methylhomoalanine, and largely rigidified proline moiety. Overall, affinity was found to increase by a factor of 1000, explained partly by an entropic advantage. All ligands adopt the same binding mode with small deviations. The residual mobility of the bound ligands is decreased across the series, and a protein side chain differs in its order/disorder behavior along with changes in the surface-water network pattern established across the newly generated protein-ligand surfaces. The enthalpy/entropy inventory displays a rather complex picture and emphasizes that thermodynamics can only be compared in terms of relative differences within a structurally similar ligand series.
- Institute of Pharmaceutical Chemistry, Philipps University Marburg, Marbacher Weg 6, 35037, Marburg, Germany.
Organizational Affiliation: