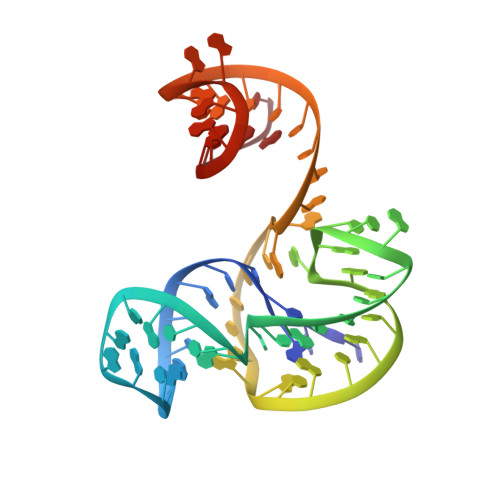
The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production.
Chapman, E.G., Costantino, D.A., Rabe, J.L., Moon, S.L., Wilusz, J., Nix, J.C., Kieft, J.S.(2014) Science 344: 307-310
- PubMed: 24744377
- DOI: https://doi.org/10.1126/science.1250897
- Primary Citation of Related Structures:
4PQV - PubMed Abstract:
Flaviviruses are emerging human pathogens and worldwide health threats. During infection, pathogenic subgenomic flaviviral RNAs (sfRNAs) are produced by resisting degradation by the 5'→3' host cell exonuclease Xrn1 through an unknown RNA structure-based mechanism. Here, we present the crystal structure of a complete Xrn1-resistant flaviviral RNA, which contains interwoven pseudoknots within a compact structure that depends on highly conserved nucleotides. The RNA's three-dimensional topology creates a ringlike conformation, with the 5' end of the resistant structure passing through the ring from one side of the fold to the other. Disruption of this structure prevents formation of sfRNA during flaviviral infection. Thus, sfRNA formation results from an RNA fold that interacts directly with Xrn1, presenting the enzyme with a structure that confounds its helicase activity.
- Department of Biochemistry and Molecular Genetics, School of Medicine, University of Colorado Denver, Aurora, CO 80045, USA.
Organizational Affiliation: