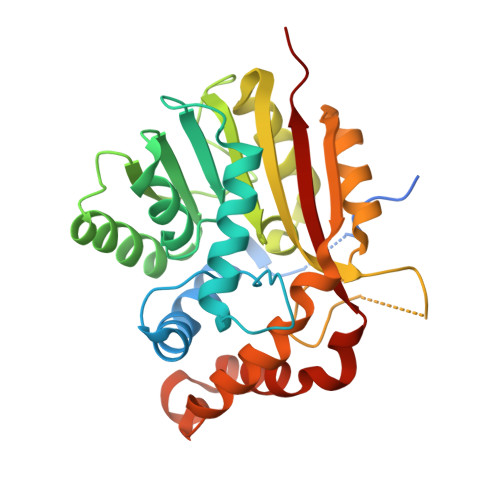
The structure of SpnF, a standalone enzyme that catalyzes [4 + 2] cycloaddition.
Fage, C.D., Isiorho, E.A., Liu, Y., Wagner, D.T., Liu, H.W., Keatinge-Clay, A.T.(2015) Nat Chem Biol 11: 256-258
- PubMed: 25730549
- DOI: https://doi.org/10.1038/nchembio.1768
- Primary Citation of Related Structures:
4PNE - PubMed Abstract:
In the biosynthetic pathway of the spinosyn insecticides, the tailoring enzyme SpnF performs a [4 + 2] cycloaddition on a 22-membered macrolactone to forge an embedded cyclohexene ring. To learn more about this reaction, which could potentially proceed through a Diels-Alder mechanism, we determined the 1.50-Å-resolution crystal structure of SpnF bound to S-adenosylhomocysteine. This sets the stage for advanced experimental and computational studies to determine the precise mechanism of SpnF-mediated cyclization.
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas, USA.
Organizational Affiliation: