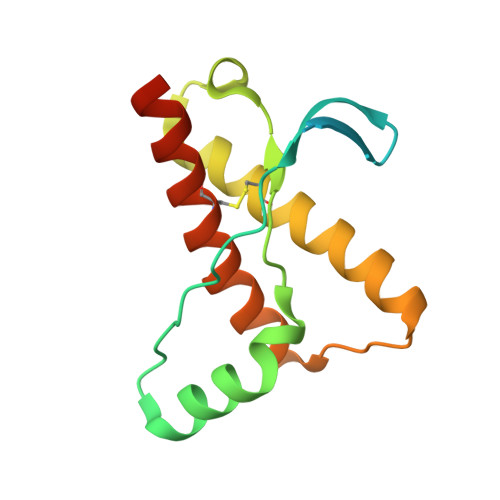
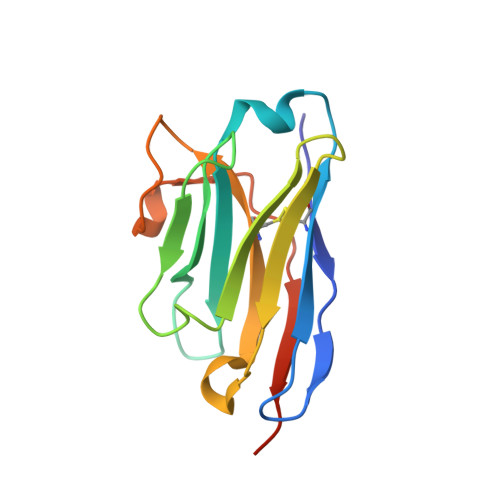
Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
Abskharon, R.N., Giachin, G., Wohlkonig, A., Soror, S.H., Pardon, E., Legname, G., Steyaert, J.(2014) J Am Chem Soc 136: 937-944
- PubMed: 24400836
- DOI: https://doi.org/10.1021/ja407527p
- Primary Citation of Related Structures:
4KML, 4N9O - PubMed Abstract:
Prions are fatal neurodegenerative transmissible agents causing several incurable illnesses in humans and animals. Prion diseases are caused by the structural conversion of the cellular prion protein, PrP(C), into its misfolded oligomeric form, known as prion or PrP(Sc). The canonical human PrP(C) (HuPrP) fold features an unstructured N-terminal part (residues 23-124) and a well-defined C-terminal globular domain (residues 125-231). Compelling evidence indicates that an evolutionary N-terminal conserved motif AGAAAAGA (residues 113-120) plays an important role in the conversion to PrP(Sc). The intrinsic flexibility of the N-terminal has hampered efforts to obtain detailed atomic information on the structural features of this palindromic region. In this study, we crystallized the full-length HuPrP in complex with a nanobody (Nb484) that inhibits prion propagation. In the complex, the prion protein is unstructured from residue 23 to 116. The palindromic motif adopts a stable and fully extended configuration to form a three-stranded antiparallel β-sheet with the β1 and β2 strands, demonstrating that the full-length HuPrP(C) can adopt a more elaborate β0-β1-α1-β2-α2-α3 structural organization than the canonical β1-α1-β2-α2-α3 prion-like fold. From this structure, it appears that the palindromic motif mediates β-enrichment in the PrP(C) monomer as one of the early events in the conversion of PrP(C) into PrP(Sc).
- Structural Biology Brussels, Vrije Universiteit Brussel , Pleinlaan 2, 1050 Brussels, Belgium.
Organizational Affiliation: