The conserved architecture of the T6SS central spike complex
Sycheva, L.V., Shneider, M.M., Basler, M., Ho, B.T., Mekalanos, J.J., Leiman, P.G.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Biological assembly 1 assigned by authors and generated by PISA (software)
Biological assembly 2 assigned by authors and generated by PISA (software)
Macromolecule Content
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| VgrG1 | 643 | Pseudomonas aeruginosa PAO1 | Mutation(s): 0 Gene Names: PA0091, vgrG1 | 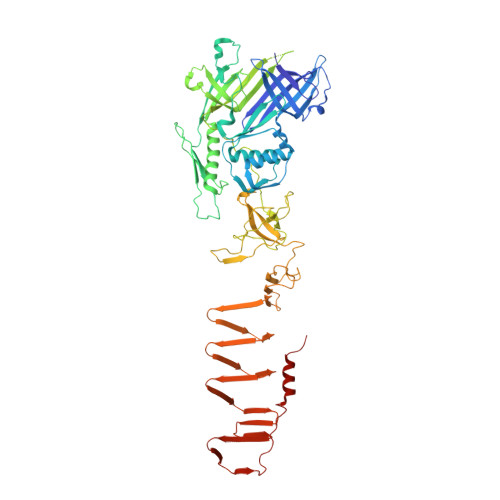 | |
UniProt | |||||
Find proteins for Q9I741 (Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)) Explore Q9I741 Go to UniProtKB: Q9I741 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9I741 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 168.275 | α = 90 |
| b = 168.275 | β = 90 |
| c = 652.849 | γ = 120 |
| Software Name | Purpose |
|---|---|
| RESOLVE | model building |
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| RESOLVE | phasing |
RCSB PDB Core Operations are funded by the U.S. National Science Foundation (DBI-2321666), the US Department of Energy (DE-SC0019749), and the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM157729. RCSB PDB uses resources of the National Energy Research Scientific Computing Center (NERSC), a Department of Energy User Facility.


