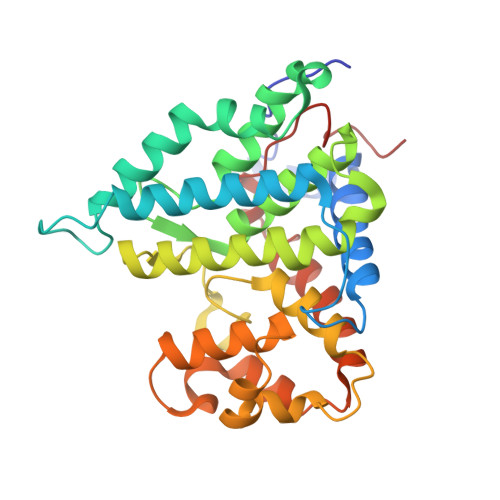
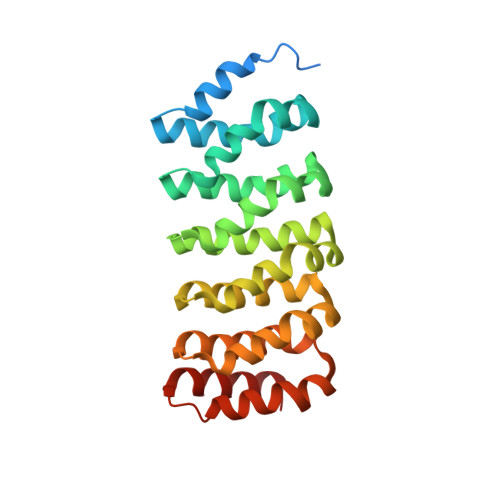
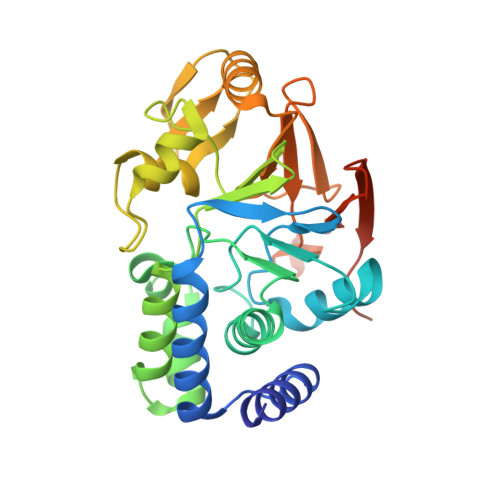
Structural basis of PP2A activation by PTPA, an ATP-dependent activation chaperone.
Guo, F., Stanevich, V., Wlodarchak, N., Sengupta, R., Jiang, L., Satyshur, K.A., Xing, Y.(2014) Cell Res 24: 190-203
- PubMed: 24100351
- DOI: https://doi.org/10.1038/cr.2013.138
- Primary Citation of Related Structures:
4LAC - PubMed Abstract:
Proper activation of protein phosphatase 2A (PP2A) catalytic subunit is central for the complex PP2A regulation and is crucial for broad aspects of cellular function. The crystal structure of PP2A bound to PP2A phosphatase activator (PTPA) and ATPγS reveals that PTPA makes broad contacts with the structural elements surrounding the PP2A active site and the adenine moiety of ATP. PTPA-binding stabilizes the protein fold of apo-PP2A required for activation, and orients ATP phosphoryl groups to bind directly to the PP2A active site. This allows ATP to modulate the metal-binding preferences of the PP2A active site and utilize the PP2A active site for ATP hydrolysis. In vitro, ATP selectively and drastically enhances binding of endogenous catalytic metal ions, which requires ATP hydrolysis and is crucial for acquisition of pSer/Thr-specific phosphatase activity. Furthermore, both PP2A- and ATP-binding are required for PTPA function in cell proliferation and survival. Our results suggest novel mechanisms of PTPA in PP2A activation with structural economy and a unique ATP-binding pocket that could potentially serve as a specific therapeutic target.
- McArdle Laboratory, Department of Oncology, University of Wisconsin at Madison, School of Medicine and Public Health, Madison, WI 53706, USA.
Organizational Affiliation: