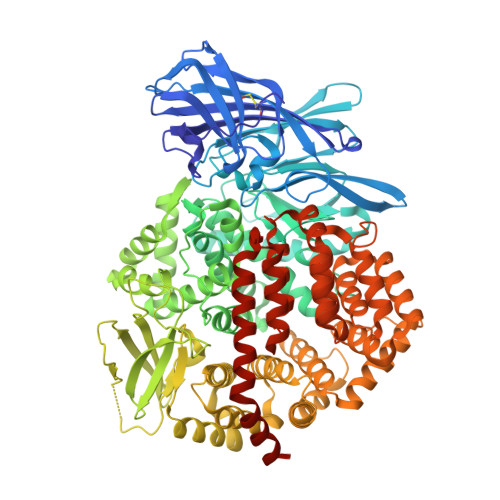
Structural insights into central hypertension regulation by human aminopeptidase a.
Yang, Y., Liu, C., Lin, Y.L., Li, F.(2013) J Biological Chem 288: 25638-25645
- PubMed: 23888046
- DOI: https://doi.org/10.1074/jbc.M113.494955
- Primary Citation of Related Structures:
4KX7, 4KX8, 4KX9, 4KXA, 4KXB, 4KXC, 4KXD - PubMed Abstract:
Hypertension is regulated through both the central and systemic renin-angiotensin systems. In the central renin-angiotensin system, zinc-dependent aminopeptidase A (APA) up-regulates blood pressure by specifically cleaving the N-terminal aspartate, but not the adjacent arginine, from angiotensin II, a process facilitated by calcium. Here, we determined the crystal structures of human APA and its complexes with different ligands and identified a calcium-binding site in the S1 pocket of APA. Without calcium, the S1 pocket can bind both acidic and basic residues through formation of salt bridges with the charged side chains. In the presence of calcium, the binding of acidic residues is enhanced as they ligate the cation, whereas the binding of basic residues is no longer favorable due to charge repulsion. Of the peptidomimetic inhibitors of APA, amastatin has higher potency than bestatin by fitting better in the S1 pocket and interacting additionally with the S3' subsite. These results explain the calcium-modulated substrate specificity of APA in central hypertension regulation and can guide the design and development of brain-targeting antihypertensive APA inhibitors.
- From the Department of Pharmacology, University of Minnesota Medical School, Minneapolis, Minnesota 55455.
Organizational Affiliation: