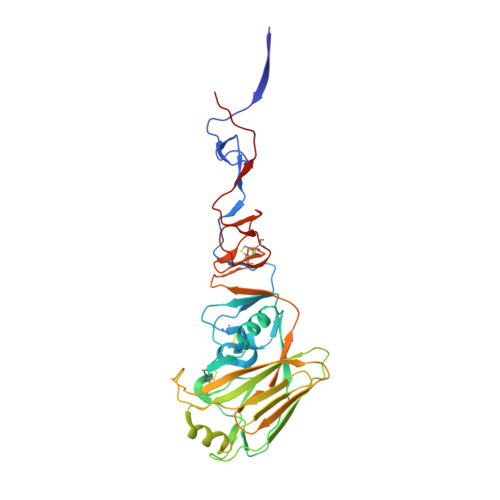
Receptor Binding by H10 Influenza Viruses.
Vachieri, S.G., Xiong, X., Collins, P.J., Walker, P.A., Martin, S.R., Haire, L.F., Zhang, Y., Mccauley, J.W., Gamblin, S.J., Skehel, J.J.(2014) Nature 511: 475
- PubMed: 24870229
- DOI: https://doi.org/10.1038/nature13443
- Primary Citation of Related Structures:
4CYV, 4CYW, 4CYZ, 4CZ0, 4D00 - PubMed Abstract:
H10N8 follows H7N9 and H5N1 as the latest in a line of avian influenza viruses that cause serious disease in humans and have become a threat to public health. Since December 2013, three human cases of H10N8 infection have been reported, two of whom are known to have died. To gather evidence relating to the epidemic potential of H10 we have determined the structure of the haemagglutinin of a previously isolated avian H10 virus and we present here results relating especially to its receptor-binding properties, as these are likely to be major determinants of virus transmissibility. Our results show, first, that the H10 virus possesses high avidity for human receptors and second, from the crystal structure of the complex formed by avian H10 haemagglutinin with human receptor, it is clear that the conformation of the bound receptor has characteristics of both the 1918 H1N1 pandemic virus and the human H7 viruses isolated from patients in 2013 (ref. 3). We conclude that avian H10N8 virus has sufficient avidity for human receptors to account for its infection of humans but that its preference for avian receptors should make avian-receptor-rich human airway mucins an effective block to widespread infection. In terms of surveillance, particular attention will be paid to the detection of mutations in the receptor-binding site of the H10 haemagglutinin that decrease its avidity for avian receptor, and could enable it to be more readily transmitted between humans.
- 1] MRC National Institute for Medical Research, The Ridgeway, Mill Hill, London NW7 1AA, UK [2].
Organizational Affiliation: