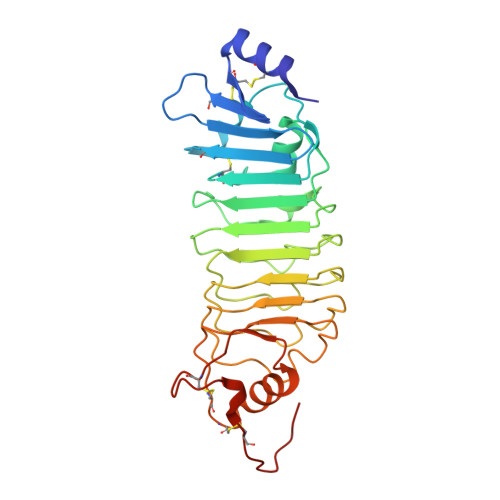
Functional Insights from the Crystal Structure of the N-Terminal Domain of the Prototypical Toll Receptor.
Gangloff, M., Arnot, C.J., Lewis, M., Gay, N.J.(2013) Structure 21: 143
- PubMed: 23245851
- DOI: https://doi.org/10.1016/j.str.2012.11.003
- Primary Citation of Related Structures:
4ARN - PubMed Abstract:
Drosophila melanogaster Toll is the founding member of an important family of pathogen-recognition receptors in humans, the Toll-like receptor (TLR) family. In contrast, the prototypical receptor is a cytokine-like receptor for Spätzle (Spz) protein and plays a dual role in both development and immunity. Here, we present the crystal structure of the N-terminal domain of the receptor that encompasses the first 201 amino acids at 2.4 Å resolution. To our knowledge, the cysteine-rich cap adopts a novel fold unique to Toll-1 orthologs in insects and that is not critical for ligand binding. However, we observed that an antibody directed against the first ten LRRs blocks Spz signaling in a Drosophila cell-based assay. Supplemented by point mutagenesis and deletion analysis, our data suggests that the region up to LRR 14 is involved in Spz binding. Comparison with mammalian TLRs reconciles previous contradictory findings about the mechanism of Toll activation.
- Department of Biochemistry, University of Cambridge, Cambridge, CB2 1GA, UK. Electronic address: mg308@cam.ac.uk.
Organizational Affiliation: