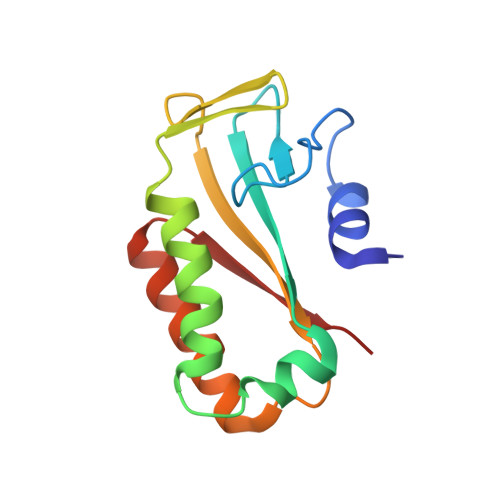
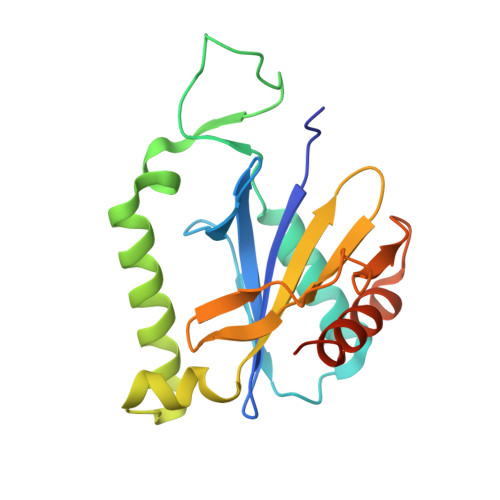
Diversification of beta-Augmentation Interactions between CDI Toxin/Immunity Proteins.
Morse, R.P., Willett, J.L., Johnson, P.M., Zheng, J., Credali, A., Iniguez, A., Nowick, J.S., Hayes, C.S., Goulding, C.W.(2015) J Mol Biology 427: 3766-3784
- PubMed: 26449640
- DOI: https://doi.org/10.1016/j.jmb.2015.09.020
- Primary Citation of Related Structures:
4ZQU, 4ZQV, 4ZQW - PubMed Abstract:
Contact-dependent growth inhibition (CDI) is a widespread mechanism of inter-bacterial competition mediated by the CdiB/CdiA family of two-partner secretion proteins. CdiA effectors carry diverse C-terminal toxin domains (CdiA-CT), which are delivered into neighboring target cells to inhibit growth. CDI(+) bacteria also produce CdiI immunity proteins that bind specifically to cognate CdiA-CT toxins and protect the cell from auto-inhibition. Here, we compare the structures of homologous CdiA-CT/CdiI complexes from Escherichia coli EC869 and Yersinia pseudotuberculosis YPIII to explore the evolution of CDI toxin/immunity protein interactions. Both complexes share an unusual β-augmentation interaction, in which the toxin domain extends a β-hairpin into the immunity protein to complete a six-stranded anti-parallel sheet. However, the specific contacts differ substantially between the two complexes. The EC869 β-hairpin interacts mainly through direct H-bond and ion-pair interactions, whereas the YPIII β-hairpin pocket contains more hydrophobic contacts and a network of bridging water molecules. In accord with these differences, we find that each CdiI protein only protects target bacteria from its cognate CdiA-CT toxin. The compact β-hairpin binding pocket within the immunity protein represents a tractable system for the rationale design of small molecules to block CdiA-CT/CdiI complex formation. We synthesized a macrocyclic peptide mimic of the β-hairpin from EC869 toxin and solved its structure in complex with cognate immunity protein. These latter studies suggest that small molecules could potentially be used to disrupt CDI toxin/immunity complexes.
- Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, CA 92697, USA.
Organizational Affiliation: